1. Introduction
The study of networks is becoming an increasingly important area of biological research. A fundamental goal in the postgenomic era is to understand cellular function in terms of complex behaviors emerging from simpler interactions at the biomolecular level. Networks offer a natural representation for aggregating biomolecular interactions for a wide range of tasks including protein-protein interaction maps, pathway diagrams and in silico based simulations. Current gene regulatory networks are reconstructed by combining data from many experiments and multiple labs using hypothesis-driven approaches, but there is a growing effort to infer biological networks from high-throughput and genome-wide data sets. This chapter discusses topological properties of gene regulatory networks, and the methods to reconstruct these networks from high-throughput data.
2. Principles in network design
Networks arise in diverse areas including telecommunications, electric grids, social interactions, and many areas of biology (for review, see Strogatz, 2001). The entire field of network analysis is beyond the scope of this work; however, some general network design principles can be considered when analyzing biological networks. We mention some the most pertinent examples here:
Shared resources: Sharing resources is often beneficial, especially in the presence of economies of scale.
Cascaded functions: Networks may be used to perform sequential sets of operations or processes that can be organized as modules that are small subnetworks.
Control and stability: Networks may exist to provide control mechanisms for an otherwise uncontrolled set of processes.
3. Defining network properties
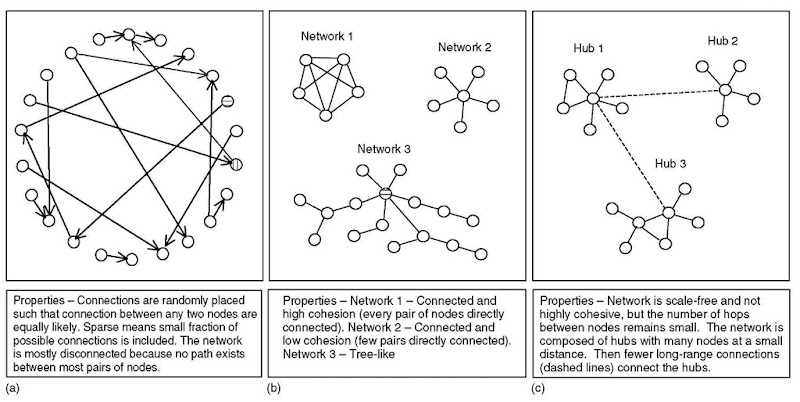
Figure 1 shows several examples of networks. Figure 1(a) illustrates a specific topology known as the Erdos-Renyi (E-R) network in which edges are randomly placed between nodes with equal probability (Erdos and Renyi, 1959). In this example, the graph is mostly disconnected so that no path exists between most pairs of nodes. One of the earliest theorems in random graph theory from Erdos and Renyi relates the number of edges in an undirected graph to the connectedness. The theorem states that the probability of the network being connected rises from 0 to 1 over the interval NlogN — cN < E < NlogN + cN, where N is the number of nodes, E is the number of edges, and c is a constant. Hence, these networks make a relatively quick transition from disconnected to connected as the number of edges approaches N logN. This example is a directed graph that is typical for a gene regulatory network in which nodes represent genes and edges represent genes influencing other genes. We can define an indegree (outdegree) as the number of edges going into (out of) each node. For example, the node with vertical crosshatches has an indegree of 1 and an outdegree of 1, whereas the node with horizontal crosshatches has an indegree of 0 and an outdegree of 1.
Several network properties are now defined. The network in Figure 1(a) is considered sparse because the number of edges is O(N) rather than O(N2) (i.e., only 16 edges out of 484 possible connections). For a directed graph with N nodes, there are N2 possible connections if a node can connect to any other node including itself. Sparseness has been observed for many biological networks (e.g., see Yeung et al., 2002); however, the E-R graph in Figure 1(a) differs from real gene regulatory networks in many other respects. The connectedness of a graph describes the degree to which paths exist between the nodes that comprise the network. Figure 1(b) shows several networks that are connected, but with different properties. Network 1, called a clique, is considered cohesive because all nodes connect directly with all others. Here undirected edges are shown, but the properties are similar for directed edges. Network 2 is also connected but is not as cohesive because only the center node connects directly with the others. Network 3 illustrates the scale-free property that is defined by the distribution of node degrees following an approximate power law relation (i.e., P(k) & k—Y). Many biological networks are scale-free (Jeong et al., 2001; Ravasz et al., 2002) with few high-degree nodes connected to many lower-degree nodes. In contrast, an E-R graph has a nodal degree distribution that is Poisson (i.e., the probability of a node having degree k is P(k) & e—ddk/k!, where d is the average node degree).
Figure 1(c) illustrates a network that is both scale-free and small-world. Small-world refers to the property that any two nodes are connected by a relatively small number of hops (transitions across edges), as reported from many biological networks (Ravasz et al., 2002). This property is quantified by the diameter which is the maximum distance between any pair of nodes in the network, where distance is defined here as the number of hops in the shortest path between a pair of nodes. In the example shown in Figure 1(c), the scale-free property is reflected by few nodes with high degrees called hubs connected to many nodes of low degree. The hubs are then joined by connections (dashed traces) so that the radius of the network remains small. Figure 1 shows other network architectures that do not show this scale-free property. For example, network 3 in Figure 1(b) is treelike with a root node. In these networks, the diameter grows as a function of the logarithm of the number of nodes.
Figure 1 Sample networks illustrate network properties, (a) Erdos-Renyi (E-R) network; (b) three sample connected networks; and (c) scale-free and small-network is based on hubs with longer range links
4. Gene regulatory networks
Much of the previous discussion can be applied to a wide variety of networks. We now get more specific and consider gene regulatory networks in which connections represent the regulatory effect of one gene on another. For example, if gene A increases (or decreases) the expression of gene B, then an arrow with a positive (or negative) sign is placed from node A to node B. The actual mechanism is that gene A is transcribed and translated into a protein which in turn increases (or decreases) the rate of transcription of gene B. The expression of gene B can have further downstream actions on other genes or produce a protein that may have other effects (e.g., an enzyme could be produced to alter metabolic pathways (Reed et al., 2003) or repair DNA (Ronen et al., 2002)). Often, multiple genes can affect the target gene, and a gene may regulate itself, (autoregulation).
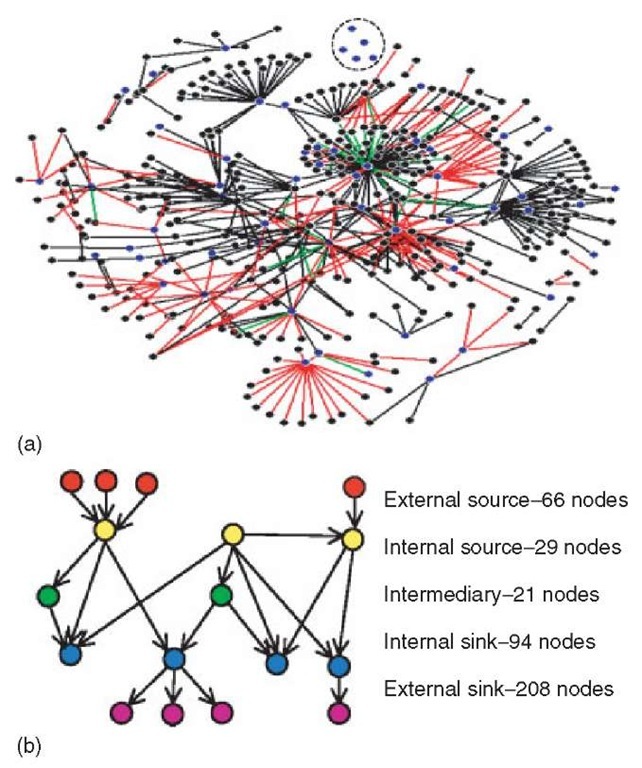
The transcriptional network from Escherichia coli is probably the most completely characterized gene regulatory network to date. Considerable knowledge of the regulatory mechanisms exists in the literature and has been condensed in databases such as Regulon DB (Salgado etal., 2000). In addition, considerable work has been done to analyze and reconstruct this network based on high-throughput data sources, as discussed later. By compiling the interactions in Regulon DB and other sources, networks similar to that shown in Figure 2(a) can be reconstructed (Shen-Orr et al., 2002). The network has 423 nodes and 578 connections. The representation does not show the autoregulation that exists in 59 of the 423 nodes. In addition, the regulatory network has three types of edges to represent activating (335), repressing (214), and both activating and repressing (29).
Several features of the network can be described using concepts already discussed. The network has a small number of nodes with high outdegree and a large number of nodes with only a single input or output connection to the rest of the network (see the starlike nodes in the center of Figure 2(a)). The node with the highest outdegree connects to 72 other nodes, and 15 other nodes have outdegree in the range of 10-26. The large number of nodes with high outdegree does not follow the Poisson distribution of E-R networks or the power law distributions of scale-free networks. Roughly, three-quarters of the nodes in the network (328 of 423) belong to a large connected component (here, a connection is assumed if any edge exists between a pair of nodes). The remaining nodes exist in components of 12 or fewer nodes. Figure 2(b) illustrates five distinct types of nodes in this network. External sources (red) and sinks (purple) are defined as having only one edge, either input or output, respectively. If one conceptualizes a flow of information from top to bottom, these nodes are starting and terminating points. Moving inward, external sources (yellow) and sinks (blue) connect to internal sources and sinks, respectively, that have two or more edges. The intermediary class (green) has relatively few nodes that connect only to internal sinks and sources (see Article 108, Functional networks in mammalian cells, Volume 6 and Article 113, Metabolic dynamics in cells viewed as multilayered, distributed, mass-energy-information networks, Volume 6.
Figure 2 The transcriptional regulation network from E. coli (Adapted from Shen-Orr SS, Milo R, Mangan S and Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genetics, 31, 64-68 by permission of Nature Publishing Group). (a) The network has 423 nodes and 578 edges and is displayed using Pajek (Batagelj and Mrvar, 2003). The regulatory network has three types of edges to represent activating (black), repressing (red), and both activating and repressing (green) connections. The 59 nodes with autoregulation are shown in blue, and the others are shown in black. (b) All but the five isolated nodes (in the dashed circle) can be placed in five classes of nodes, as labeled, based on the number and types of edges (see text for details)
5. Motif-based analysis
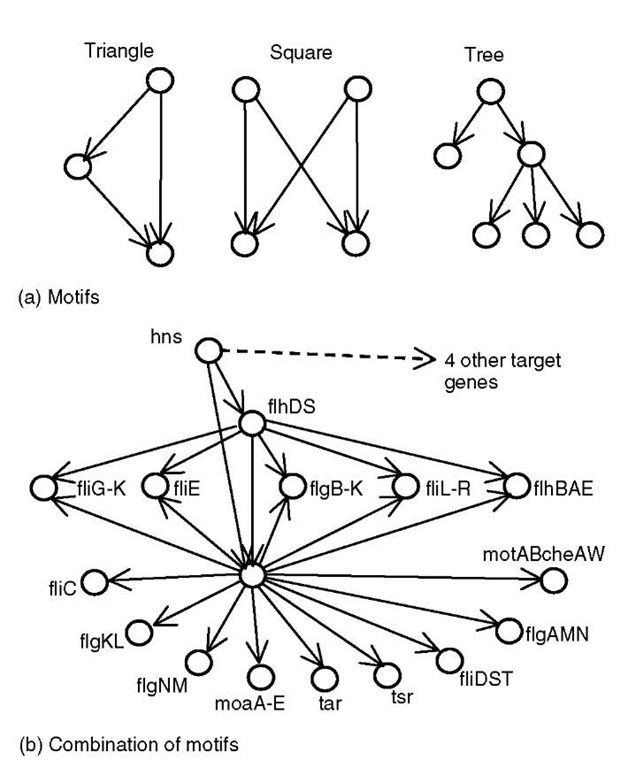
The connectivity of networks is only one static aspect of the interactions in the cellular environment. Yet, the complexity contained at this level of description is already considerable. Sufficiently large networks have interesting large-scale properties, such as power law degree distributions and relatively small diameter. Another level of analysis seeks to identify local properties of the network such as motifs. A motif is a recurring, possibly inexact, pattern in the network that may represent a reused module within the gene regulatory network. Moreover, the number of occurrences of motifs is usually compared to what is expected by chance, that is, the number discovered in randomized networks of similar properties (e.g., networks with randomized connections that preserve the in- and outdegree of each node). With this kind of analysis, motifs such as those shown in Figure 3(a) are found (Shen-Orr etal., 2002; Milo etal., 2002; Kershenbaum etal., 2003). The feed-forward triangle and the squares (also called bifan (Milo etal., 2002)) occurred at roughly two and three times the rate, respectively, as for the randomized network (Z-scores equal 4.9 and 7.5, respectively, Kershenbaum et al., 2003). The two examples are simple motifs that can be rigidly defined; however, motifs with more flexible structures can also be discovered. The descendent tree is a source node with several children, perhaps spread across levels (a two-level descendent tree is called a single input module in Shen-Orr et al., 2002).
Figure 3 Samples of motifs discovered in the transcriptional regulation network from E. coli in Figure 2. Simple motifs in (a) can be combined in larger structures such as the example in (b) that corresponds to regulatory modules to assemble the flagella (see text for details)
One might also analyze networks as a composition of simple motifs. For example, the composite of motifs in Figure 3(b) is a composition of a descendent tree and several feed-forward triangles (Kershenbaum et al., 2003). The genes in this subnetwork are mostly associated with regulating the sequence and timing of proteins required to assemble the flagella (Kalir et al., 2001) (compare Figure 3(b) with Figure 1 in the previous reference). Interestingly, the ancestor in this motif is a gene called H-NS, a master regulator in maintaining bacterial homeostasis under rapidly changing environments (Schroder and Wagner, 2002). Directly downstream of H-NS is flhDC, the master controller for the flagellar assembly process characterized in the previous work cited (Kalir et al., 2001). Note that the composite motif in Figure 3(b) was discovered on the basis of topology alone without regard to the name or function of the nodes. Hence, these results suggest that motif-based analyses can point to functional modules without full characterization of the nodes.
6. Biological interpretation of E. coli network topology
The layered network structure in Figure 2(b) suggests several properties. One can envision an “information flow” from a small number of sources (internal or external) to a much larger number of external sinks that can affect cellular processes. In the middle of the network, intermediary nodes may act as information integrators that subsequently affect larger numbers of downstream events. The layered networks produce short paths, perhaps suggesting that short temporal delays are favored over additional information integration that might be achieved with more levels of processing. Moreover, the negative autoregulation that is present in over 40% of E. coli transcription factors is thought to decrease rise-times in gene expression and hence may also decrease delay (Rosenfeld et al., 2002). Note that negative feedback loops at the level of gene regulation are not found, suggesting that additional stability as provided by this mechanism is not favored. Open loop control typically has better temporal responses and may be sufficient in this case. For example, the fast rate of division and high rate of error in DNA replication are thought to allow E. coli to rapidly “adjust parameters” in the metabolic pathways to make optimal use of the nutrients available in the current medium (Rosenfeld et al., 2002). Hence, this case suggests there is a regulation at the population level instead of at the level the individual gene network in this organism.
7. Reconstruction of pathways from high-throughput data
The E. coli network considered so far is constructed as compilations of data collected from the literature and other sources. Hence, the network represents a considerable part of the existing knowledge about the system and summarizes much of the underlying biology even though the functions of some genes may be better characterized than others. As mentioned earlier, however, E. coli is a special case in that it has been the object of a sustained research effort as one of the preferred model organisms for understanding prokaryotic biology. However, the knowledge base is sparser for most higher organisms. To address these limitations, considerable effort is being made to reconstruct networks based on high-throughput data and genome-wide data (see Article 110, Reverse engineering gene regulatory networks, Volume 6 and Article 118, Data collection and analysis in systems biology, Volume 6). However, high-throughput data is no panacea, and initial attempts to reconstruct large-scale, full kinetic models of the cell have faced serious difficulties (Rice and Stolovitzky, 2004). From a theoretical perspective, the underdetermined nature of the problem (more unknowns than equations) implies that a unique solution is not generally possible because an infinite number of reconstructed systems are consistent with any given set of data. To deal with this nonuniqueness, the solution space is often limited by a priori, and often reasonable, assumptions such as linearity and sparseness, but even then the limitations of existing data render these approaches as mostly theoretical exercises except for reasonably small systems with high-quality data (Gardner et al., 2003; see also Article 110, Reverse engineering gene regulatory networks, Volume 6).
Another line of research seeks a reconstruction of the connectivity without full kinetic detail. We shall mention several efforts that are characteristic of the challenges facing the field (for a comprehensive review, see van Someren et al., 2002). A common approach is Bayesian inference methods as first used by Friedman et al. (2000) to analyze gene expression data. While these methods can handle noisy and incomplete data, the initial results showed that even three-node networks were hard to reconstruct in the yeast galactose metabolic pathway (Hartemink et al., 2001); however, much of the trouble may lie in the quality of the experimental data and not the method per se. Later work by the same group showed better results using synthetic gene networks where the researchers had better control of the data quality and quantity (Smith et al., 2003). In an elegant work (Gardner et al., 2003), researchers were able to reconstruct much of a nine-gene subnetwork in a DNA repair pathway in E. coli by controlled perturbations of a subset of the member genes. This work made the assumption of sparseness of the pathway connections and included a robust experimental design that kept low levels of noise in the real-time PCR measurement of transcript levels. In other recent studies (Segal et al., 2003; Troyanskaya et al., 2003), genome-wide yeast expression data and preliminary clustering were used to determine likely functional modules, that is, the sets of genes working together to perform a particular function. In addition, other data sources (candidate regulatory genes (Segal et al., 2003) and yeast two-hybrid data (Troyanskaya et al., 2003)) were combined to predict the functional modules. Hence, better results are found for understanding the regulation of sets of genes (versus single genes in initial attempts), more and perhaps better-quality data, and the use of complementary data types in addition to expression data alone.
8. Reconstruction of biological networks with pair-wise conditional correlation with a perturbed gene
We have proposed an alternative reconstruction method that seeks to determine the network connectivity only without generating a predictive model of the system (Rice et al., 2004). Starting with topologies based on the network in Figure 2(a), the network is endowed with dynamic behavior based on the work of Yeung et al. (2002). We propose an experimental design in which a single node is perturbed and the resulting effect on the rest of the network is measured. By computing the Pearson correlation of the expression profile of the perturbed gene to all the other genes in the network, the functionally connected genes can be inferred when the correlation is above a set threshold. The process is repeated in a gene-by-gene fashion in order the reconstruct as large a network as needed. With this method, the network can be reconstructed with a high degree of accuracy that produces a reasonably low normalized fraction of false-positives and false-negatives.
The low reconstruction error rates of this and other similar studies are encouraging enough that full-scale automated reconstructions of gene regulatory networks may be possible in the foreseeable future with genome-wide data sets. False connections may be problematic for large network reconstruction (e.g., the E. coli 423 node network has ~4232 possible connections so that a 1% false-positive error translates into more that 1000 false-positives, twice as many as the number of real connections). Therefore, reconstruction methods should usually be complemented with false-positive reducing approaches. Specifically, an optimal threshold to reject connections can be determined by modeling the distribution of correlation values of separate components representing connected versus disconnected nodes (Rice et al., 2004). Additional heuristics were developed to distinguish directly connected genes from indirectly connected genes that may also show high correlations (i.e., X ^ Z is distinguished from X ^ Y ^ Z).
While a large sparse network may make false-positives problematic, experimental noise may increase the rate of false-negatives. Other aspects of the E. coli gene regulatory network facilitated reconstruction. Specifically, the network hierarchical shape with large descendent trees that link relatively few sources to many sinks appears of be a topology that favors reconstruction. Specifically, the case of a gene being regulated by only one other (such as external sinks) is easier to reconstruct because the input and output show high correlation. In contrast, the infrequent structure of many nodes impinging on a single node is more difficult to reconstruct as the output is a function of many inputs and hence shows weaker correlation. Indeed, methods have shown similar difference in reconstruction accuracy for the case of single versus many inputs to a node (Smith et al., 2003). This latter study has shown difficulty with feedback pathways, but these are not seen to exist in the currently known E. coli network in Figure 2(a).
9. Toward a theory of biological systems
While allowing an unprecedented view into complex biological systems, the recent explosion of information has also opened a “Pandora’s box”. The systems biologist is like Newton under the tree who is pondering the apple in his hand. Fortunately, Newton connects the dots between the apple, the Earth, and his headache and develops calculus as the analytical tool to both understand and predict all motions, terrestrial and otherwise. A similar grand challenge for the systems biologist is to create a framework that allows the biologist to unify existing knowledge, to compare biological systems across species, and to make quantitative predictions of experimental manipulations. Network approaches are emerging as important ingredients in this quest. Networks are natural tools for compiling and mining high-throughput, genome-wide datasets. Networks are also tools to aggregate simpler interactions into large systems with complex behaviors. At the intersections of these tasks, large datasets can be used to infer network connectivity, pathways, and potentially the kinetics between the interacting cellular players. Going the other direction, network-based tools such as motif discovery may help to decompose large systems into modules to more easily deduce functional relations. Clearly, networks will play important roles as the systems biologist tries to reign in the complexity and put the lid back on Pandora’s box.