1. Introduction
Cellular function arises from the interaction of thousands of distinct chemical components, including DNA, RNAs, proteins, and small molecules. Determining the networks of interactions between such molecules has become a major interest of the biological community, with the mapping of chemical interactions underway in many model organisms. This effort involves the consolidation of data from focused biochemical, molecular biology, and genetics studies (Bhalla and Iyengar, 1999; Davidson etal., 2002), with data from high-throughput, systematic approaches. Here, we discuss approaches for building network models from biological data, a task referred to as network inference or reverse engineering (see Article 109, Analyzing and reconstructing gene regulatory networks, Volume 6 and Article 60, Extracting networks from expression data, Volume 7) (D’haeseleer etal., 2000; Banerjee and Zhang, 2002; Brazhnik etal., 2002; de Jong, 2002; Wyrick and Young, 2002; Li and Wang, 2003; Stark etal., 2003a,b; Friedman, 2004; Herrgard et al., 2004; Rice and Stolovitzky, 2004; Taverner et al., 2004).
Network inference approaches can be categorized (Table 1) by the level of physical detail in the model, the biological data used to construct the model, the inference method or underlying mathematical structure, and the type of biological insight desired from the model. There is no direct correspondence between the features in these different categories. For instance, different types of biological data and different model types can be used to infer a qualitative network model to describe cellular function. In the next sections, we will discuss the different levels of physical detail used in molecular network models and the types of biological data that are available for network inference. We will then describe four recent studies in detail (Beer and Tavazoie, 2004; Liao et al., 2003; Ronen et al., 2002; Gardner etal., 2003) to present different strategies for inferring gene regulatory networks. These four studies are notable in their emphasis on gaining biological insight, and each uses a different type of biological data to accomplish the inference task. We will also discuss the novel advances and limitations of each approach. In the final section, we will provide concluding remarks and suggest future directions for network inference studies.
Table 1 Distinguishing between network modeling approaches
| Distinguishing characteristic | Examples |
| Level of description (Figure 1) | Topological, qualitative, and quantitative models |
| Data used to construct model | Physical and functional interactions sequence, physical interaction, and molecular abundance data; time-series and steady state |
| perturbation experiments | |
| Model type | Boolean, Bayesian, linear, and nonlinear models |
| Model purpose | Descriptive, interpretive, and predictive models |
2. Cellular network descriptions
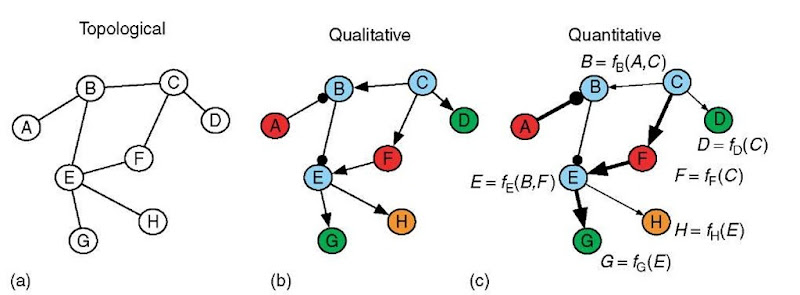
There are different levels of detail (Rice and Stolovitzky, 2004) with which one can describe the structure of a molecular network (Table 1; Figure 1). Each level of description is appropriate for answering a different set of biological questions, and successful modeling involves a choice of detail appropriate for the question posed. In the first, and most simplified model (Figure 1a), the network is described as a set of homogenous components interacting through topological connections, indicating only which components interact. Models of this type have been used to compare the structure of molecular networks with other interaction networks, such as social networks made up of human components, and information networks consisting of website components. The potential biological insight from this work includes determining the universal principles responsible for the evolution, organization, and function of molecular networks (reviewed in Barabasi and Oltvai, 2004).
The second, and most predominant type of model (Figure 1b) includes additional details to describe the functional heterogeneity of the components and the nature of the interactions. These qualitative connections indicate properties such as the direction (causality) and sign (activating or inhibiting) of an interaction. This type of model is most prevalent because the function of a molecular network is often obvious based on its qualitative structure and biological intuition.
The third, and most detailed network model (Figure 1c) provides a quantitative description of the interactions between network components, indicating how each component behaves as a function of its inputs. This level of detail is required to predict functions that are not conceptually intuitive, such as determining the mode of action of a pharmaceutical compound or simulating dynamic cellular behavior. With increasing model detail, the task of inferring the structure of a network becomes more challenging and requires more biological data.
The level of detail of network models can also differ by the amount of physical realism described by the model (Table 1). A network connection can represent the interaction of two components that physically interact, as in the case of protein-protein interaction networks (Uetz etal., 2000; Ito etal., 2001; Gavin et al., 2002; Ho et al., 2002; Giot et al., 2003; Li et al., 2004), or it can simply represent a functional relationship between two components. For example, a functional connection between two genes in a gene regulatory network means that a change in abundance of the product (mRNA or protein) of one gene affects the abundance of the product of the other gene. The physical interactions that mediate such a connection are unknown or ignored for the purpose of a simplified model (Brazhnik etal., 2002). This review focuses on the modeling of gene regulatory networks, so all of the connections described are functional in nature.
Figure 1 Molecular interaction network models can be described at different levels of detail. (a) Topological models describe graphs with homogenous molecular components and topological connections, indicating only which components interact. (b) Qualitative models include details of the functional heterogeneity of the molecular components (indicated by component color) and contain information on the direction (causality) and sign (activating or inhibiting) of interactions. Arrows begin at the causal factor and end at the component regulated by that factor. Arrow-tip terminated connections are activating; circle-tip connections are inhibiting. Component colors indicate different molecule functions; for instance, red components might be transcription factor proteins and blue components might be kinase proteins. (c) Quantitative models indicate how each component behaves as a function of the state of its inputs. For instance, component B will respond as a function of its inputs from components A and C. The strengths (indicated by line width of the arrow) of the interactions are captured in such a model
3. Inferring gene networks from biological data
The achievable level of model detail and corresponding network inference strategy both depend on the type of biological data that is available. For example, data on the absolute or relative abundance of molecular species can provide a quantitative measure of the response of molecular networks to stimuli. This can enable topological, qualitative, or quantitative network inference, and is especially important for qualitative and quantitative models. Technologies such as gene expression microarrays (Schena etal., 1995; Lockhart etal., 1996), which monitor mRNA abundance, have made the systematic collection of such molecular abundance data accessible to individual laboratories.
The most common approach to gaining biological insight with systematically collected molecular abundance data is to cluster genes into groups with similar patterns of expression over a set of conditions or a time course following a stimulus (Eisen et al., 1998; Wen et al., 1998; Alon et al., 1999; Tamayo et al., 1999; Holter et al., 2000). Coexpression clustering has been reviewed elsewhere (see Article 90, Microarrays: an overview, Volume 4) (Brazma and Vilo, 2000; D’haeseleer et al., 2000; Gerstein and Jansen, 2000; Lockhart and Winzeler, 2000; Quackenbush, 2001). Groups of genes that share expression patterns have been shown to function in similar cellular processes, and their protein products are more likely to participate in physical interactions with one another (Eisen et al., 1998; Ge et al., 2001; Jansen et al., 2002). Observing correlated expression for a set of genes is useful for the construction of a gene regulatory network because the coexpressed genes often have a similar set of regulatory inputs. However, correlation alone is insufficient to uncover the regulatory inputs and construct a model of causal gene-gene interactions. Such relationships can be identified using additional data types and/or methods that control the causal factor in the experimental design.
Data suitable for inference of causal gene relationships include DNA sequence data (Tavazoie et al., 1999; Bussemaker et al., 2001; Pilpel et al., 2001; Wang et al., 2002; Segal et al., 2003c; Beer and Tavazoie, 2004; Haverty et al., 2004), annotated lists of regulators (Ihmels et al., 2002; Segal et al., 2003a; Qian et al., 2003; Haverty et al., 2004), physical protein-DNA binding data (Bar-Joseph et al., 2003; Liao et al., 2003; Gao et al., 2004; Kao et al., 2004), time-series experiments (Arkin and Ross, 1995; Arkin et al., 1997; Holter et al., 2001; Ronen et al., 2002; Perrin et al., 2003; Sontag et al., 2004), and targeted steady state perturbation experiments (Kyoda et al., 2000; Ideker et al., 2001; Pe’er et al., 2001; Wagner, 2001; Bruggeman et al., 2002; de la Fuente et al., 2002; Kholodenko et al., 2002; Wagner, 2002; Wang et al., 2002; Yeung et al., 2002; Tegner et al., 2003; Gardner et al., 2003; Sontag et al., 2004; Vlad et al., 2004). Methods that use this data to uncover the causal relationships include Bayesian inference, signal decomposition, and parameter estimation, and will be described below.
In the remainder of this section, we will focus on four examples of how molecular network inference has been used to gain biological insight. Each of the four studies uses a different type of biological data to accomplish the network inference task. Our emphasis will be on the novel advances and limitations of each approach, and the lessons in network modeling that can be drawn in each case.
4. Inference from sequence data
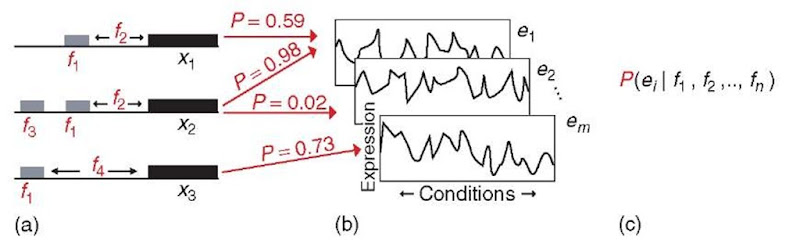
A recent study by Beer and Tavazoie (2004) used systematically collected mRNA abundance data in conjunction with DNA sequence data to study the combinatorial regulatory program underlying gene expression (Figure 2). As described above, genes that have correlated expression patterns are likely regulated by one or more common transcription factor proteins. The response of a gene to the simultaneous binding of more than one transcription factor may not be a simple linear superposition of the responses to the individual factors (Davidson et al., 2002; Zeitlinger et al., 2003). Beer and Tavazoie (2004) hypothesized that in addition to the combination of transcription factors regulating a particular gene (incoming network connections), the binding location and orientation of these factors relative to each other and to the start of protein translation would influence the expression of that gene. Information about the binding location and orientation of transcription factors is encoded in the DNA sequence, in which short patterns of nucleotides termed cis-regulatory motifs are recognized and bound by transcription factor proteins (Bulyk, 2003; Wasserman and Sandelin, 2004).
Figure 2 Approach taken by Beer and Tavazoie (2004) to determine quantitative connections between sequence determinants and gene expression. Red features are learned using the approach. (a) Sequence determinants of gene expression are enumerated as features, f, and include the presence of a motif, its orientation, and its distance from the translation start site. (b) Expression profiles, ei, of genes under different conditions. The sequence determinants in (a) are mapped to corresponding gene expression profiles using a probabilistic function. (c) The conditional probability of a gene exhibiting a particular expression profile, ei, based on sequence features, fi, in its promoter. This probability is determined for all the expression profiles and sequence features using a Bayesian network
To accomplish their task, Beer and Tavazoie (2004) employed the Bayesian framework to describe the probabilistic dependencies between DNA sequence elements and gene expression profiles. First, they obtained data showing mRNA abundance changes in response to different environmental conditions, giving an expression profile for each gene. Next, they clustered the genes into sets of similar expression profiles, ei (Figure 2b). They examined the promoter regions upstream of each gene, xt, within a cluster of coexpressed genes for shared cis-regulatory motifs. Each motif was assigned a feature number, fi, that could be used to indicate the presence (1) or absence (0) of that motif for a particular gene, xi (Figure 2a). The motif orientation and distance from the translation start site were also assigned feature numbers. Finally, they described a Bayesian network (Pearl, 1988; Friedman etal., 2000; Pe’er etal., 2001; Perrin etal., 2003; Segal etal., 2003a,b,c; Friedman, 2004) mapping DNA sequence features (fi, f2,… , fn), to gene expression patterns (e,) through the conditional probability, P(et|f1, f2,… , fn), that a gene with a particular set of sequence features will participate in expression pattern i (Figure 2c). To train their model, Beer and Tavazoie (2004) searched through the space of sequence features to find a network (N) with the maximum probability of being correct given the observed data (D), using Baye’s rule: P(NID) = P(N)P(D|N)/P(D). They withheld some data from the training set to test the predictive power of their model.
Beer and Tavazoie (2004) demonstrated their approach with gene expression data from the yeast Saccharomyces cerevisiae, responding to environmental stresses (Gasch etal., 2000) and a cell-cycle time course (Spellman etal., 1998). One expression profile they observed showed the stress-induced change in abundance of gene products involved in ribosomal RNA transcription and processing. They identified two cis-regulatory motifs, termed PAC and RRPE (Sudarsanam et al., 2002), that are overrepresented in the promoter regions of these genes. Their Bayesian network model showed that the presence of the RRPE motif within 240 bp of the translation start site (ATG) of a gene indicates a 22% probability that the gene will exhibit the expression profile. Similarly, the presence of the PAC motif within 140 bp of the ATG indicates a 67% probability of the same. Importantly, the presence of both motifs within those respective locations is 100% indicative that the gene will exhibit the expression profile. Beer and Tavazoie (2004) describe this combinatorial program as AND logic, which strictly speaking means that the pattern is followed if both features are present and not followed if either individual feature or neither feature is present. Whether that same logical function would describe the response of a gene to the presence of activated versions of transcription factors that bind the PAC and RRPE motifs remains to be seen.
Beer and Tavazoie (2004) identified many other combinatorial sequence determinants of gene expression patterns in yeast, including some resembling OR and NOT logic functions. They demonstrated that those sequence features are predictive of the mRNA abundance changes of greater than 70% of the yeast genes responding to the observed environmental stress conditions. They also identified a program of combinatorial transcriptional regulation controlling the embryonic and larval development of the nematode Caenorhabditis elegans. They reasonably suggested that more and higher-quality expression data will enable the identification of many more regulatory programs.
The use of a Bayesian network approach provided some key benefits in this study. It allowed easy incorporation of heterogeneous determinants of gene expression, including not only sequence but also motif orientation and location. In addition, combinatorial gene regulation is commonly considered to involve a nonlinear response of gene expression to multiple simultaneous inputs. The Bayesian framework provides a probabilistic model with which these nonlinearities can be explored. Interestingly, it appears that regulation of genes by the PAC and RRPE motifs cannot be described using pure AND logic. For instance, genes containing only PAC or only RRPE can still exhibit the observed expression pattern (see above). Under these circumstances, which may be common in biological systems, it is likely that a linear model could also capture much of the important behavior. The additional benefit of using sequence data is that influential sequence determinants that are not associated with the binding of a particular protein, such as GC content affecting the physical properties of DNA, may influence transcription and could be learned using approaches similar to that of Beer and Tavazoie (2004).
Despite the excellent promise of this approach, some limitations should be considered in the future development of network inference strategies. For example, it is not yet clear how much of the information on the regulation of a gene is encoded in the DNA sequence in its promoter region. In higher organisms, the boundaries of promoter regions are less clear, and sequence determinants are commonly located tens of thousands of nucleotides upstream of a gene (Alberts, 2002). To infer a gene regulatory network model, one would also like to know the transcription factors that bind a given cis-regulatory motif. Coupling the approach of Beer and Tavazoie (2004) with genome-wide protein-DNA binding data determined using chromatin immunoprecipitation (Lee et al., 2002) is one strategy to consider. Finally, as Beer and Tavazoie (2004) discuss, their statistical approach of averaging over genes that follow a common expression pattern may not enable the learning of very complex combinatorial regulation programs that have few or unique instances in a genome. However, the latter limitation may be partially overcome by using a strategy of comparing sequences of similar organisms (Bulyk, 2003; Cliften etal., 2003; Kellis etal., 2003; Wasserman and Sandelin, 2004).
5. Inference from physical interaction data
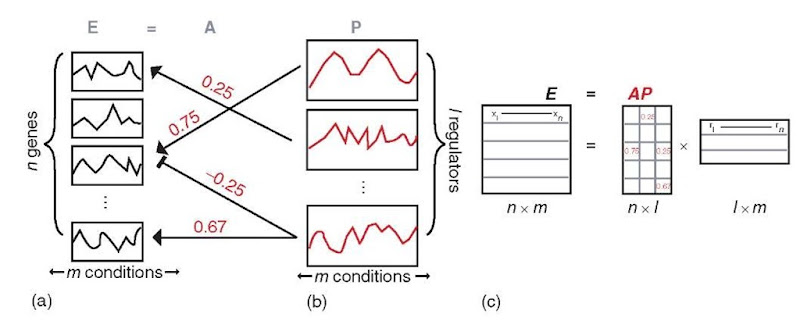
A recent study by Liao et al. (2003) demonstrated how mRNA abundance data and known qualitative network interactions from physical protein-DNA binding data (Ren et al., 2000; Lieb et al., 2001; Iyer et al., 2001; Lee et al., 2002; Kurdistani et al., 2002) can be used to determine quantitative gene regulatory network connections (Figure 3). Their approach also enabled the discovery of the activity profiles of a set of transcriptional regulators over a time course and a set of environmental conditions. The aim of their approach was to model the behavior of each of the regulated genes in the network as a linear combination of the activity profiles of the transcription factors that regulate that gene. The approach of Liao et al. (2003) is termed network component analysis (NCA), and is suggested to yield more biologically relevant linear combinations than other signal decomposition approaches because of its use of prior information on regulatory relationships.
The network model includes a layer of transcription factors (Figure 3b) interacting with a larger set of regulated genes (Figure 3a). The NCA is provided with a matrix, E, containing experimental data monitoring the abundance of each gene product in the network over a time course or set of conditions (Figure 3a). The approach by Liao et al. (2003) performs a linear decomposition of the expression data, E, into a connectivity matrix, A, and regulator profile matrix, P (Figure 3c). The connectivity matrix, A, describes the interactions directed from the transcription factors to the regulated genes. The A matrix is a pruned model of connectivity, representing a minimal nonoverlapping set of connections between transcription factors and regulated genes. Feedback connections are not allowed. The nonzero elements of matrix A are known from the physical protein-DNA binding studies, and the sign and magnitude (weights) of the elements (regulatory connections) are determined using NCA (Figure 3). The transcription factor activity (TFA) profile matrix, P, is determined using NCA, and describes the activity of the transcription factors (Figure 3b) over the same time course or conditions as the regulated genes. Decomposition of the expression data is achieved using a least-squares minimization, min \\E — AP||2, through an iterative procedure starting with the known network structure and random connectivity strengths.
Figure 3 Approach taken by Liao et al. (2003) to determine quantitative gene regulatory connections and transcription factor activity (TFA) profiles using molecular abundance data and physical protein-DNA binding data. Red features are learned using the approach. (a) The algorithm is supplied with a matrix of experimental data, E, monitoring the abundance of n genes over m time points or environmental conditions. (b) The TFA profiles of the l transcription factors are contained in the P matrix. These TFA profiles are learned using the approach. The transcription factors direct regulatory connections, contained in matrix A, toward the genes in the network. The structure of these connections is known a priori, but the connection strengths and signs are learned using the approach. (c) The expression profiles, E, of the n regulated genes in the network are a linear combination of the l transcription factor activity profiles, P, weighted by the connectivity matrix, A
Liao et al. (2003) demonstrated the NCA approach using mRNA abundance data taken over the time course of the yeast cell cycle (Spellman et al., 1998) and physical protein-DNA binding data obtained using systematic chromatin immuno-precipitation (Lee et al., 2002). They focused on 11 transcription factors known to regulate gene expression in a cell-cycle-dependent manner. To satisfy the criteria for NCA, the network of regulated genes was reduced to 441 genes under the control of these 11 factors and 22 other transcription factors, for a total of 33 regulators. Liao et al. (2003) used the NCA to determine the connectivity strengths and signs between these 33 transcription factors and the 441 genes, as well as the activity profiles of the transcription factors over the course of the cell cycle. Interestingly, they observed that the regulator activity profiles captured by the NCA do not necessarily correspond to the molecular abundance profiles of those regulators. They observed cases in which the activity profile of the regulator protein exhibited cyclical behavior, even though its corresponding mRNA abundance did not.
Even more compelling is a similar observation made by Liao and colleagues in a second study examining the response of Escherichia coli to carbon source transition (Kao et al., 2004). They decomposed the behavior of 100 genes responding to a shift from glucose to acetate growth media, into regulatory contributions by 16 transcription factors and the associated activity profiles of those regulators. One of the transcription factors they examined was CRP (combinatorial regulation programs), which requires the binding of the small molecule, cAMP, for its regulatory activity. Kao et al. (2004) showed that the activity profile (TFA) of the CRP-cAMP complex determined by the NCA algorithm matches the abundance profile of cAMP. With the NCA approach, it will be possible in principle to identify transcription factors whose abundance profile differs from their activity profile. These transcription factors would likely be regulated by means other than molecular abundance changes, including regulation by phosphorylation, localization, or complexation.
A limitation of this approach is the dependence on prior knowledge of the semiqualitative network connections. However, the construction of such network models is becoming more feasible with the use of DNA sequence data (described above) and the determination of physical protein-DNA binding locations using chromatin immunoprecipitation.
6. Inference from time-series experiments
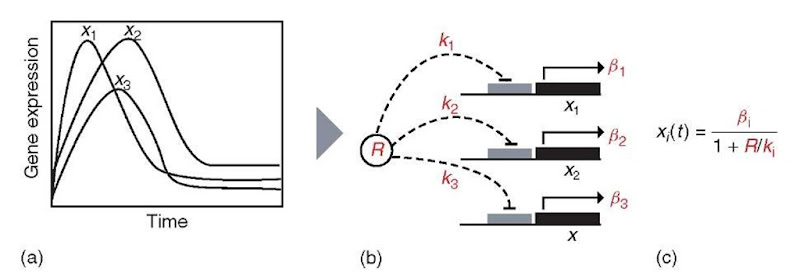
A recent study by Ronen etal. (2002) used time series of molecular abundance measurements to determine kinetic parameters (quantitative connections) for a network with previously known qualitative (direction and sign) connections (Figure 4). Their goal was to demonstrate the inference of a more complete descriptive model that could be used to predict the dynamics of the entire network on the basis of the observation of a single gene. The molecular network they examined was the DNA damage response and repair (SOS) pathway in E. coli. This network regulates the cellular response to DNA damage and involves more than 100 genes. Under normal conditions, the LexA transcriptional repressor (“R” in Figure 4b) blocks expression of other genes in the network. Under DNA damaging conditions, the RecA protein becomes activated and mediates LexA degradation, thereby relieving the repression of the SOS network genes.
Ronen et al. (2002) measured the temporal change in abundance (Figure 4a) of eight genes in the SOS network, including lexA and recA, following UV irradiation. They estimated the abundance of the proteins in the network using a fluorescent reporter protein placed under the control of the eight promoters. Their goal was to fit kinetic parameters for the production rate from each derepressed promoter (Pi), and the effective affinity of the LexA protein for each promoter (kt), based on the time-dependent LexA repressor activity (R(t); Figure 4b) and the observed promoter activity with respect to time (X (t); Figure 4a). First, the LexA repressor activity (R(t)) was determined on the basis of singular value decomposition (SVD) from the different experiments. Next, the kinetic parameters were estimated using a Michaelis -Menten model for the regulatory dynamics (Figure 4c). This equation is specific to the system they investigated, and would take a different form if any of the genes were activated by LexA, or if LexA bound cooperatively to any of the promoters.
Figure 4 Approach taken by Ronen et al. (2002) to determine kinetic parameters (quantitative connections) for gene expression using time-series measurements of molecular abundance, and a prior model of the qualitative connections. Red features are learned using the approach. (a) Time-varying molecular abundances measured for every regulated species. (b) Regulatory interactions show that gene expression, xi, is dependent on the repressor activity, R, the binding affinity for the repressor, ki, and the expression level in the absence of repressor, pi. (c) Michaelis-Menten model, showing the relationship between gene expression and the parameters in (b)
Ronen et al. (2002) were able to estimate kinetic parameters that reproduced the observed time-dependent upregulation of eight SOS network genes upon simulated LexA degradation. They discussed how the temporal order of the alleviation and reapplication of repression for the eight genes, as determined by the magnitude of the repressor binding affinity parameters, k, agrees with the order in which the genes function in the DNA repair process. Others have observed similar mechanistically significant temporal ordering of gene expression in different systems (Spellman et al., 1998; Laub et al., 2000; Kalir et al., 2001; Zaslaver et al., 2004), and this appears to be a key feature in transcriptional network dynamics. Ronen et al. (2002) also demonstrated how their kinetic model could be used to estimate the relative abundance of the active LexA regulator over the time course of the experiments, showing agreement with experiments that directly determined protein abundance (Sassanfar and Roberts, 1990). They also showed that the calculated error in estimated parameters could be used to anticipate additional regulation for a particular promoter that is not captured on the basis of LexA activity alone.
There are some limitations of this approach as a general solution for determining quantitative connections in gene regulatory networks. The strategy taken by Ronen et al. (2002) is specific for modeling the response of a group of genes to changes in activity of a single regulatory factor. The approach cannot describe the time-dependent response of a group of genes to multiple varying transcription inputs, or under circumstances in which there is feedback between the regulated genes and the regulatory factor. Elucidating the behavior in these more complicated regulatory system would require knowledge of the cis-regulatory logic describing multiple simultaneous inputs (Beer and Tavazoie, 2004). Another limitation of this approach is that it requires prior knowledge of the qualitative connections in the network, because the selection of an appropriate kinetic model will depend on the system structure. For the most part, the elucidation of such a qualitative network model remains a challenge. In principle, time-series data could be used to obtain the causality information required for inferring a network of qualitative connections with no prior structural information (Arkin and Ross, 1995; Arkin et al., 1997; Holter et al., 2001; Sontag et al., 2004). For example, Perrin et al. (2003) used the time-series data collected by Ronen et al. (2002) to infer a network of qualitative interactions between the eight genes in the SOS subnetwork.
7. Inference from perturbation experiments
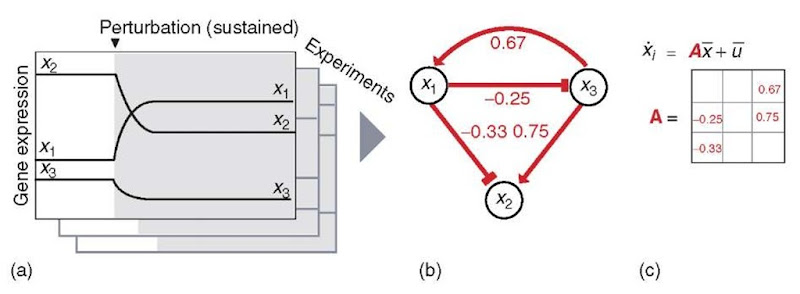
A recent study by Gardner et al. (2003) demonstrated the use of molecular abundance data measured in response to targeted steady state perturbations to infer a gene network model with quantitative connections (Figure 5). The resulting network model describes the quantitative influence of each gene on the expression of every other gene in the network. One of the goals of the study was to use the resulting model to identify the major regulators in the network, which in their model need not be transcription factor proteins. Another goal was to predict the targets of unknown perturbations (for instance, the molecular target of a pharmaceutical compound).
Figure 5 Approach taken by Gardner et al. (2003) to determine quantitative connections between genes on the basis of molecular abundance measurements following targeted steady state perturbations. No prior information on network structure was required. Red features are learned using the approach. (a) Steady state molecular abundance measured for every species following genetic perturbations. (b) Quantitative network model describes the influence of each gene on every other gene. The connectivity matrix, A, encodes these influence weights. (c) Linear model for the accumulation of each species, Xi, based on the abundance of the other species, x, the connectivity matrix, A, and the perturbations, u. At steady state, the molecular abundances are not changing (Xi = 0)
In the approach taken by Gardner et al. (2003), a cellular system near steady state is subjected to the specific perturbation (overexpression or downregulation) of a gene thought to have an influence on a network of interest. Once the system returns to steady state, the response of the system to the perturbation is measured by determining the molecular abundance of every species in the network (Figure 5a). Near a steady state, the behavior of the system can be approximated by a linear model (Figure 5c) describing the rate of accumulation, Xi, of each species, i, in the network in terms of the abundance of every gene product in the network, X, the quantitative connectivity matrix, A, and the perturbation, u, made to the system. At steady state, the abundance of each species is not changing (X, = 0), so the equation in Figure 5(c) reduces to AX = -u. After making many perturbation-response measurements, the quantitative coefficients of A (Figure 5b), which represent the influence of each gene on every other gene, can be determined using linear regression.
Gardner etal. (2003) tested their approach, termed network identification via multiple regression (NIR), on the SOS network in E. coli (described above). As a starting point, they applied the NIR method to a nine-gene subset at the core of the network. The authors used plasmid-borne copies of each gene to individually alter its abundance in nine separate experiments and measured the resulting changes in mRNA abundance for all nine components (Figure 5a). The NIR method was able to correctly identify 25 of the previously identified regulatory relationships between the nine genes, as well as 14 relationships that may be novel interactions or false positives. Moreover, the network model obtained by the NIR algorithm correctly identified the recA and lexA genes, the known principal regulators of the SOS response, as having the strongest influence (largest regulatory weights) on the other genes in the network. Thus, the model can be used to identify which genes should be perturbed to elicit a maximal response from the network.
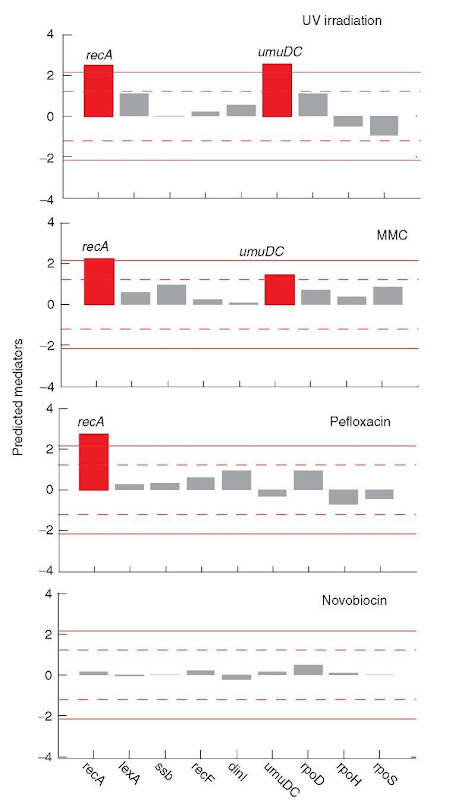
The network model obtained by the NIR algorithm was also used to identify the genes that mediate the network response to a particular stimulus. As illustrated in Figure 6, the network model correctly identifies the recA gene as the key mediator of the SOS network response to treatment with UV radiation, mitomycin C (MMC), and the quinolone antibiotic pefloxacin (each of which causes DNA damage). For novobiocin (Figure 6), a quinolone that does not cause DNA damage, recA is not predicted as the mediator of the expression response. The predictive power of the network model obtained using the NIR algorithm is due to its use of quantitative connections, and this level of model detail was specifically chosen to enable the identification of compound mode of action.
There are some limitations of the NIR approach for inferring molecular network structure and function. The predictive capability of the model was gained at the expense of its ability to describe many of the molecular interactions with mechanistic detail. For instance, while the NIR approach can include regulatory connections directed by nontranscription factors, the physical interactions that mediate those connections are often unknown, making it difficult to interpret the meaning of any particular connection. Other limitations of the approach include the challenge of delivering targeted (single gene) perturbations, and the difficulty in knowing which genes to perturb to most efficiently reconstruct the network model.
8. Concluding remarks and future directions
We have examined in detail four recent approaches to gene network inference (Beer and Tavazoie, 2004; Liao et al., 2003; Kao et al., 2004; Ronen et al., 2002; Gardner et al., 2003). These four approaches can be distinguished by several different model characteristics (Table 1). Despite differing in their underlying model structure, each of the four approaches provides a gene network model with some quantitative detail about the connections. For instance, the Bayesian network model of Beer and Tavazoie (2004) described the behavior of a cluster of genes as a probabilistic function of DNA sequence determinants, and the linear influence network model of Gardner et al. (2003) described the accumulation of one species as a weighted sum of the abundance of the other species in the network.
Although every gene regulatory network model has connections that represent functional relationships rather than physical interactions, the models can differ in their descriptive power. For instance, the approaches of Beer and Tavazoie (2004), Liao et al. (2003), and Ronen et al. (2002) describe connections between transcription factors (or the corresponding DNA sequence motifs to which they bind) and regulated genes. These interactions occur through well-understood mechanisms, and identifying such an interaction is therefore descriptive of the molecular behavior. The approach by Gardner et al. (2003), on the other hand, yields connections that are often mediated by several unknown physical interactions, making descriptive interpretation challenging. The resulting model, however, enables the prediction of behaviors resulting from the perturbation of nontranscription factors, including protein targets of pharmaceutical compounds.
Figure 6 Prediction of genes mediating response to four different stimuli. The network model identified with the approach of Gardner et al. (2003) (Figure 5) was used to predict the mediators of expression responses following UV irradiation and treatment with three drug compounds. In the case of treatment with UV radiation, mitomycin C (MMC), and pefloxacin, all of which cause DNA damage, the recA gene is correctly predicted as the mediator of the expression response. For treatment with novobiocin, which does not damage DNA, recA is not predicted as the mediator of the expression response. Lines denote significance levels: P = 0.3 (dashed), P = 0.1 (solid)
The current trend in molecular network inference is in coupling different types of experimental data to improve the quality and completeness of the inferred model. The Bayesian formalism has been invoked in many studies as the most convenient approach for including previous knowledge and tolerating missing information (Beaumont and Rannala, 2004; Friedman, 2004). As Liao et al. (2003) and Kao et al. (2004) demonstrated, it is also possible to include prior information on network structure in linear models using a constraint-based approach (see Article 112, Constraint-based modeling of metabolomic systems, Volume 6). Another type of data that has not been discussed here are the synthetical lethal interactions, determined systematically in yeast by Tong etal. (2004). These interactions represent functional relationships between genes that are involved in similar or related cellular processes, and coupling these data with network inference approaches should prove fruitful. Finally, the use of physical protein-protein interaction data (Uetz etal., 2000; Ito etal., 2001; Gavin etal., 2002; Ho etal., 2002; Giot etal., 2003; Li etal., 2004) and metabolic network structure information (Covert et al., 2001, 2004) to trace the physical interaction pathways responsible for the functional relationships between genes will help elucidate gene network structure and extend the value of gene regulatory networks as descriptive models of cellular function.
Research into approaches for network inference would benefit from the availability of large, standardized sets of molecular abundance data collected with the goal of network inference in mind. As we have described in this study, time-series experiments and targeted steady state perturbation experiments will be particularly useful. The availability of such data in several model organisms (such as E. coli, S. cerevisiae, and C. elegans) would significantly advance biological discovery.