Introduction
In 1989, at the same time that the first amplified fragment length polymorphism (AFLP) markers were first described using the polymerase chain reaction (PCR), a new class of repeats was also being reported. The large tandem repeat (LTR) polymorphisms had been referred to as minisattellite polymorphisms. The new class of di-, tri-, tetra- and pentanucleotide repeats were referred to as micro-sattelites. These markers became known as short tandem repeats (STRs). The amplification of LTR AFLP markers in general appeared to the relatively error free, with stutter bands or copying errors (extra bands in descending numbers of repeats) relatively infrequent. In actuality they do occur if a sensitive enough detection technique is used. In contrast, the newly discovered dinucleotide repeats were difficult to use because of the frequent occurrence of stutter bands. Three, four and five nucleotide repeat AFLPs have a decreasing frequency of stutter bands. The use of high base pair STR AFLP loci made it practical to use them for human gene mapping, parentage testing and identity purposes. As more of the markers became available, it was possible to choose those that were more polymorphic as well as those with different size fragments to begin making highly discriminating multiplexes. The use of fluorescent detection and automated equipment, originally for the sequencing of DNA, made it possible to create fluorescent multiplexes with automated detection. Unfortunately, it was some time before these discoveries could be moved into routine use. For purposes of simplicity the term ‘STR’ will be used to refer to any STR AFLP typing system.
Early History of Forensic STR Kits
Forensic and paternity testing laboratories became very interested in the possibility of using STRs for routine use. Many laboratories tried to develop their own multiplexes and detection systems. However, a patent was issued to Dr Caskey and the University of Texas, who in turn licensed all STR technology to the Promega Corporation, limiting the amount of outside work on the development of multiplexes. The widespread use of standardized kits had to wait for the Promega Corporation and PE Biosystems to release kits.
The first kits released by the Promega Corporation included single locus kits for the loci CSF1PO, F13A01, F13B, FESFPS, HPRTB, LPL, THO1, TPOX and vWA (incorrect nomenclature, should be VWA03, similarly to F13A01) and multiplex kits that could be detected using silver staining, including the CTT multiplex (CSF1PO, TPOX and THO1) and the FFV (F13A01, FESFPS, VWA03). Subsequently, a silver STR III system was released with the loci D16S539, D7S820 and D13S317. Figure 1 (and see Table 1) is a silver-stained gel with CTT on the right side and FFV on the left side of the gel.
The next evolution was the introduction of fluorescently labeled primer multiplexes. The nature of fluorescence and fluorescent detection is described below. With the introduction and wider availability of devices capable of reading fluorescent gels, fluorescent STR technology became much more practical. The first device commercially available for visualizing multicolored fluorescent images was ABI Applied Biosystems model 373 sequencing device which used ABI’s four-color sequencing technology (see below). (Note: Perkin Elmer Corporation subsequently acquired ABI and recently PE Applied Biosystems separated from Perkin Elmer. Products from this company will be referred to as PE Biosystems.) Subsequently, Molecular Dynamics, Hitachi, Pharmacia and others introduced devices that could visualize at least two colors. The ability to detect PCR STR multiplexes fluorescently simplified the detection from silver staining, and meant that multiplexes in at least two different colors could be detected, allowing for larger numbers of loci to combine. It also meant that machines could do the detection with minimal hands-on time compared with a detection procedure such as silver staining. From a forensic standpoint this means that, if the amount of DNA is limited, a single amplification could produce results on as many as nine or ten loci, generating population frequencies comparable with those obtained from restriction fragment length polymorphism (RFLP) testing with much less expenditure of time and staff. (See below for supporting information.)
The first generation of commercially available fluorescent multiplexes was produced by Promega and PE Biosystems. The first two fluorescent multiplexes released by Promega were labeled with fluoroscein (Table 2) and contained the loci CSF1PO,TPOX, THO1 and VWA03 (CTTV) in the first and F13A01, FESFPS, F13B and LPL (FFFL) (Table 3)in the second. PE Biosystems released two multiplexes, one labeled with the dye FAM called ‘Green’ and containing THO1, TPOX, CSF1PO and the sex identifier Amelogenin, and a second one labeled with JOE (Table 4) called ‘Blue’ and containing D3S1358, VWA03 and FGA. Subsequently, Promega released a third fluorescein-labeled quadplex called GammaSTR Multiplex that contained the loci D16S539, D7S820, D13S317 and D5S818 (Table 3).
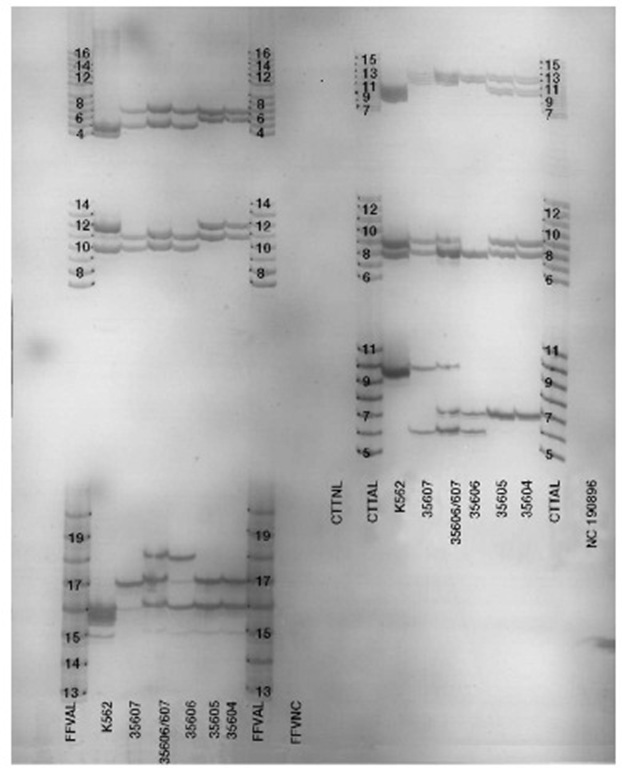
Figure 1 Silver-stained gel with both Promega CTT (right half of gel) and FFV (left half of the gel).CTTAL and FFVAL indicate the allele ladders.The lanes are: outside allele ladders; negative amplification control; positive amplification control (K562 cell line); 35607 (alleged father); mixture of 35607 and 35606; 35606 (child), and 35605 and 35604 (the mother).The results of the tests are presented in Table 1. The loci from top to bottom are CSF1PO, TPOX and TH01 on the right and F13A01, FESFPS, VWA03 on the left.Note: on the right in TPOX, where there is a mixture of 35606 and 35607, 35607 is type 8,9, 35606 is type 8, when the two are mixed the 8 band is much darker. The DNA is separated under conditions that make the DNA become single-stranded, forming doublets that can be seen at some of the loci.The VWA03 locus tends to form more pronounced stutter (repeat number minus one repeat bands) than the others; these are noticeable in the ‘positive control’ and other samples.At the VWA03 locus the child has an allele (*18) not found in the mother or the alleged father. This confirms an exclusion found previously PE Biosystems came out with a three-color kit called Profiler, which consisted of Green 1, Blue and a new multiplex called ‘Yellow,’ containing the dye TAMRA and the loci D5S818, D13S317 and D7S820 (Table 3).
In England the Forensic Science Service were also developing their own multiplexes to test evidence and profile criminals who had been convicted of significant crimes. The Royal Canadian Mounted Police (RCMP) in Canada were also looking at different multiplexes. Some of these multiplexes shared loci with those available in the United States; some were different.
Table 1 Results of silver stained CTTand FFV found in Figure 1.
| Negative | Positive | 35607 | 35607/6 | 35606 | 35605 | 35604 | |
| Locus | Control | Control | AF | Mixed | Child | Mother | Mother |
| CSF1PO | NR | 9,10 | 11,12 | 11,12 | 12 | 10,12 | 10,12 |
| TPOX | NR | 8,9 | 8,9 | 8,(9) | 8 | 8,9 | 8,9 |
| THO1 | NR | 9.3 | 6,10 | 6,7,10 | 6,7 | 7 | 7 |
| F13A01 | NR | 4,5 | 5,7 | 5,7 | 5,7 | 6,7 | 6,7 |
| FESFPS | NR | 10,12 | 10,11 | 10,11 | 10,11 | 11,12 | 11,12 |
| VWA03 | NR | 16 | 17 | 16,17,18 | 16,18 | 16,17 | 16,17 |
Table 2 Fluorescent dyes used to label primers for fluorescent STR detection
| Common name | Chemical name | Abs | Em | User |
| Fluorescein | Fluorescein | 494 | 518 | Promega |
| 5-FAM | 5-Carboxyfluorescein | 494 (493) | 517 (522) | PE Biosystems (Blue) |
| JOE | Carboxyrhodamine | 520 (528) | 548 (554) | PE Biosystems (Green) |
| TAMRA | Carboxytetramethylrhodamine | 542 | 568 | PE Biosystems (Yellow, old) |
| NED | Unknown | (553) | (575) | PE Biosystems (Yellow, new |
| TMR | Tetramethylrhodamine | 555 | 580 | Promega |
| ROX | Carboxy-X-rhodamine | 570 (587) | 590 (607) | PE Biosystems (Red) |
| CRX | Carboxy-X-rhodamine | 570 | 590 | Promega |
Table 3 STR loci in Promega’s fluorescent multiplex kits
| STR locus | Powerplex 1.1/1.2 | Powerplex2.1/2.2 | Powerplex 16.2 | CTTV | FFFL | Gamma |
| D16S539 | + (fl) | + (prop) | + (fl) | |||
| D7S820 | + (fl) | + (prop) | + (fl) | |||
| D13S317 | + (fl) | + (prop) | + (fl) | |||
| D5S818 | + (fl) | + (prop) | + (fl) | |||
| CSF1PO | + (TMR) | + (prop) | + (fl) | |||
| TPOX | + (TMR) | + (TMR) | + (TMR) | + (fl) | ||
| AMEL | + (TMR) | + (TMR) | + (TMR) | |||
| THO1 | + (TMR) | + (fl) | + (fl) | + (fl) | ||
| VWA | + (TMR) | + (TMR) | + (TMR) | + (fl) | ||
| D18S51 | + (fl) | + (fl) | ||||
| D21S11 | + (fl) | + (fl) | ||||
| D3S1358 | + (fl) | + (fl) | ||||
| FGA | + (TMR) | + (TMR) | ||||
| D8S1179 | + (TMR) | + (TMR) | ||||
| Penta D | + (prop) | |||||
| Penta E | + (fl) | + (fl) | ||||
| F13A01 | + (fl) | |||||
| FESFPS | + (fl) | |||||
| F13B | + (fl) | |||||
| LPL | + (fl) |
Table 4 STR loci combined in each PE Applied Biosystems AmpF STR kit
| STR locus | Dye color | Profiler | Profiler Plus | Cofiler | Blue | Green 1 |
| D3S1358 | Blue | + | + | + | + | |
| VWA03 | Blue | + | + | |||
| FGA | Blue | + | + | + | ||
| Amel | Green | + | + | + | + | |
| THO1 | Green | + | + | + | ||
| TPOX | Green | + | + | + | ||
| CSF1PO | Green | + | + | + | ||
| D5S818 | Yellow | + | + | |||
| D13S317 | Yellow | + | + | |||
| D7S820 | Yellow | + | + | + | ||
| D8S1179 | Green | + | ||||
| D21S11 | Green | + | ||||
| D18S51 | Green | + | ||||
| D16S539 | Blue | + |
At this point, with the large number of STR loci, a demand for a standardized panel in the United States and a need for there to be at least some sharing of loci with forensic counterparts in Canada, England and Europe, the Technical Working Group on DNA Analysis Methods (TWGDAM) implemented a multi-laboratory evaluation of those STR loci available in kits in the United States. The loci chosen would be the PCR-based core of a national sex offender file required under the 1994 DNA Identification Act. The national program was initially called Combined DNA Indexing System or CODIS for short.
The TWGDAM/CODIS loci were announced at the Promega DNA Identification Symposium in the fall of 1997 and at the American Academy of Forensic Sciences meeting in February 1998. The following loci were chosen to be part of what was originally called the CODIS 13 loci: CSF1PO, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D21S11, FGA, THO1, TPOX and VWA03. These loci overlapped with the Forensic Science Services multiplexes and the Interpol multiplexes. The thirteen loci can be obtained in two amplifications using Profiler Plus and Cofiler (Table 3) from PE Biosystems, in two amplifications using Powerplex 1 and Powerplex 2 from Promega (Table 2), or in a single reaction with Powerplex 16, when it is released by Promega (Table 2).
Fluorescent Dyes
Before discussing the equipment used to detect fluorescent STRs, some understanding of fluorescent dyes is necessary. Fluorescent dyes or minerals when subjected to light at one wavelength, such as ultraviolet (UV) light or black light, will give off colored light at a slightly different wavelength or color. A characteristic of fluorescent dyes or materials is that the compound is excited at one frequency of light, referred to as its absorption or excitation peak, and emits or gives off light at a different frequency, referred to as its emission peak.
Ethidium bromide when bound to double-stranded DNA and exposed to a UV light source at 365 nm gives off pink light, whereas SYBER green gives off a green light. Ethidium bromide is about one-tenth as sensitive as SYBR green when using 365 nm UV light. This may be due to the fact that the peak absorption of ethidium bromide is 518 nm, with a peak emission at 605 nm (seen as pink to red light). The tail on the excitation side is quite broad so that there is some excitation at 365 nm. In contrast, the absorption peak for SYBR green is 494 nm, with an emission peak at 521 nm (seen as yellow to green light). This is much closer to the frequency of the light source, and therefore more able to use the energy coming out of the light source. Many different kinds of fluorescent probes that have uses other than labeling DNA have been described.
To label a PCR primer with a fluorescent dye, the dye is attached to the 5′ end of the molecule. Since DNA is translated from 5′ to 3′, it is at the very end of the primer and should not affect the amplification or binding of the primer if made correctly. One of the oldest fluorescent dyes is fluorescein. Many devices have been made that will detect fluorescein, and it has been used extensively to label antibodies and other materials. Many of these dyes have been used for a long time and are in the public domain. Others have been developed for specific projects. The dyes used by PE Biosystems were originally proprietary and part of a patented four-color DNA sequencing system (Blue, Green, Yellow and Red). These dyes are now becoming more readily available. The dyes used commercially in the fluorescent detection of PCR-based STR systems are listed in Table 4, with the available information. The PE Biosystem Red (ROX) dye and CRX have not been mentioned. They are normally used for an internal size standard (see STR Detection, below).
Fluorescence Detection Equipment
The equipment used to detect the product of fluores-cently labeled STR tests falls into two categories. The first are devices that scan a gel after it has been electrophoresed. Examples of this are the Hitachi FMBIO Fluorescent Scanner, the Molecular Dynamics Fluorimager and the Beckman Genomics SC scanner. The Hitachi and Molecular Dynamics scanners use a laser as a light, with filters to identify the proper frequency and a charge coupled device (CCD) camera to capture the image. The Beckman Genomics SC incorporates a monochromatic xenon light source and uses filters to detect the appropriate light for the CCD camera. The CCD camera scans back and forth over the gel as it is exposed to the light source and detects the various fluorescent colors using filters that change. This type of equipment has flexibility because different formats of electrophor-esis gels can be used and scanned. The output is in the form of an electronic image with bands that look much like a set of RFLP bands. Figure 2A is an example of actual images (light on dark) recorded by the CCD camera, and Figure 2B is the reverse image (dark on light) that is reminiscent of an RFLP autoradiograph or lumigraph. It should be noted that the scanning time is added on to the electrophoresis time, with increased time for each color read.
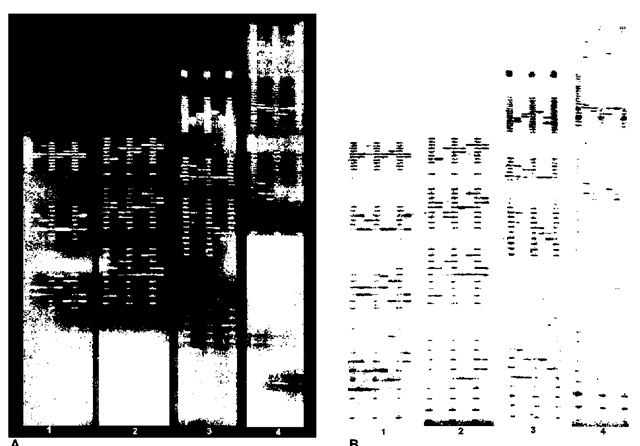
Figure 2 (A) The original CCD images of Powerplex 1.1 (panels 1 and 2) and Powerplex 2.1 (panels 3 and 4). (B) The reverse image of the same images.Panel 1 contains the TMR loci from top to bottom CSF1PO, TPOX, TH01 and VWA03.Panel 2 contains the fluorescein loci from top to bottom D16S539, D7S820, D13S317 and D5S818.Panel 3 contains the TMR loci from top to bottom FGA, TPOX, D8S1179 and VWA03.Panel 4 contains the fluorescein loci from top to bottom: Penta E, D18S51, D21S11,TH01 and D3S1358.The same samples are loaded between the ladders.The sample format is the TWGDAM format when an internal size standard is not used.The overlapping loci for sample confirmation are TH01, TPOX and VWA03.
The second type of imaging system is a real-time system, in which the DNA fragments, after the bands have been resolved, pass beneath a light source scanner that recovers the spectrum of light from the different fluorophors. This is the ABI Prism system from PE Biosystems. It includes the older model 373 DNA Sequencer and 377 DNA Sequencer, which use slab acrylamide electrophoresis to separate the DNA fragments, and the 310 Genetic Analyzer, which uses capillary electrophoresis to separate the DNA fragments. Capillary electrophoresis is a technology in which a fine glass capillary is filled with a proprietary separation polymer. The sample is pulled into the capillary by applying an electric current to it. Using high voltage electrophoresis (12 000 V), the DNA fragments are then separated over the length of the column and move past a laser detector. The 377 system can put approximately 60 samples on a gel at one time, and with modifications 96. In contrast, the 310 CE system does one sample at a time, with a separation time of approximately 20 min. However, as this is automated, a cassette can be filled with samples for testing and left to run unattended. The output of these devices is not a CCD image but a series of electropherograms with a profile for each color scanned (nominally Blue, Green, Yellow and Red). Since these are difficult to interpret, the computer software provides decomposed single color graphs. An example of Profiler Plus and Cofiler for a single sample is presented in Fig. 3. Figure 4 contains an electropherogram of the single amplification Pro-mega PowerPlex 16.2 System run on an ABI PRISM 310 Genetic Analyzer.
One of the extremely useful characteristics of fluorescent imaging devices is that the amount of light read by the detection device is quantified. The electropherograms produced by the real-time scanners is quantitative. However, the CCD image can also be used as a scanning densitometer to determine the amount of light in each peak. Since there is one fluorescent molecule per band, the amount of fluorescence is linear with the number of molecules in a band. This allows for many different types of analysis to be performed on the data generated. One of the more important of these is the ability to detect mixtures (see below).
PCR Setup and Detection
The manufacturers of the kits have done forensic validations of the kits, however each laboratory is responsible for the individualized validation required before testing. The guidelines for those validations for laboratories in the United States are governed by the DNA Advisory Board Guidelines (as of 1 October 1998), as implemented by ASCLD-LAB. Other areas of the world are regulated by other guidelines, unless they are also ASCLD-LAB accredited.
Virtually all of the procedures governing sample handling, contamination control and setup for the PCR apply to STR AFLP testing. Many of the procedures for STR typing are the same as those for DQA1, PM and D1S80 typing. The extraction procedure, quantitation, concentration (if needed) and setup are similar, if not identical. The reaction volumes for the two commercial STR kits vary. Promega’s reaction is a 25 ul volume and limits the maximum sample to 10 ul, whereas PE Biosystems uses a 50 ul reaction volume and can take as much as 20 ulof sample. It may therefore be necessary to concentrate samples when using the Promega system. Amplification conditions are those stated in the manufacturers’ protocols. The only major differences between DQA1, PM and D1S80 occur in the detection of the amplified product.
STR Detection
The major difference in the typing of the STR loci is the ability to include an internal size standard if the detection device used has multicolor capability. Under the TWGDAM guidelines forensic samples are to be placed adjacent to an allele ladder, as seen in Fig. 2. As the Beckman Genomics SC only has two filters (fluor-escein and tetramethylrhodamine), an internal ladder could not be used, so the adjacent ladder format is used. In this situation there is no special preparation for detection. If the four-color Hitachi FMBIO II Fluorescent Scanner, or ABI Prism 377 or 310, and an internal standard are used the detection setup is modified slightly. As part of the set-up a ROX ladder for PE Biosystems kits (Fig. 3), or a CRX 60-500 bp ladder (Fig. 4) or a CRX 60-400 (Fig. 5) used with Promega kits, is added to all samples including the allelic size ladders. The internal size standard is used to size all fragments within a lane by detector supplied software and assigns repeat numbers from the allele ladder sizings.
Use of Internal Size Standards:
It was previously demonstrated that within gel variation in DNA migration could be compensated for by placing a size ladder within the lane and measuring each fragment with the internal size standard. This allows for highly precise measurements of fragments. Since the electrophoresis systems used to detect the STR loci have the capability of resolving as little as 1 bp difference, it is necessary to have precise measurements to make sure that the fragment sizes can be accurately converted to repeat numbers. This would not be critical if all STRs were regular; that is, always have four base pairs for the tetranucleotide STRs. However, this is not the case. In THO1, one of the more common alleles, especially in Europe, is the THOl 9.3 allele. Examples of this are seen in Fig. 1 (the positive control) and in Fig. 2B, panel 1, third sample from the left, in the third locus from the top. For other loci, such as FGA or D21S11, there are also intermediate repeats (see below).
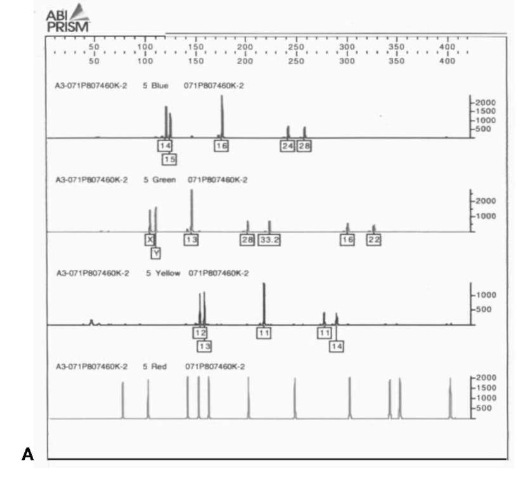
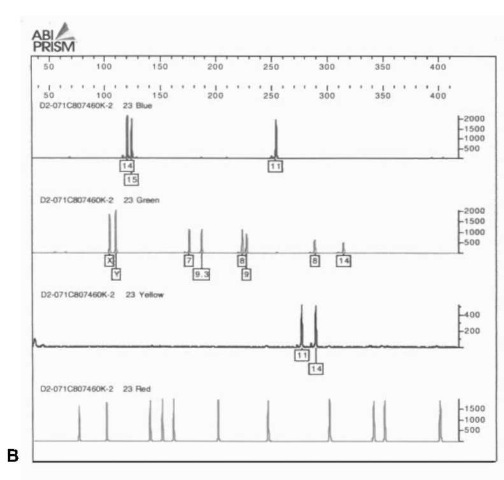
Figure 3 (A) Profiler Plus decomposed electropherogram.From left to right the loci are: Blue D3S1358, VWA03 and FGA; Green Amelogenin, D8S1179, D21S11 and D18S51; Yellow D5S818, D13S317 and D7S820.Red is the ROX sizing ladder. (B) Cofiler decomposed electropherogram.From left to right the loci are: Blue D3S1358 and D16S339; Green Amelogenin, THO1, TPOX and CSF1PO; Yellow D7S820.Red is the ROX sizing ladder.The overlapping loci for sample confirmation are Blue D3S1358, Green Amelogenin and Yellow D7S820.
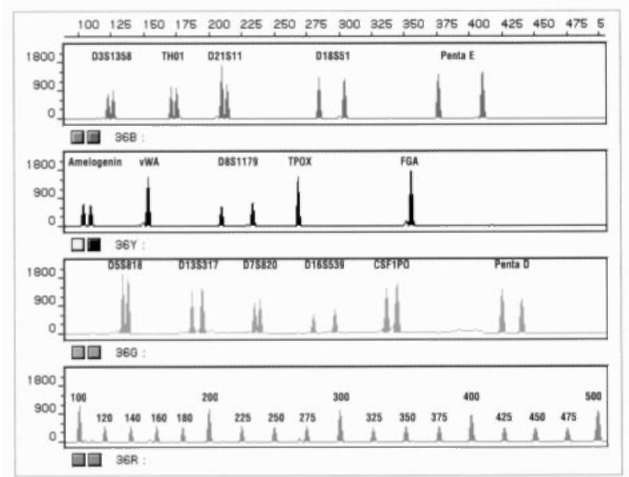
Figure 4 Electropherogram of a single DNA sample amplified using the 16-locus prototype Powerplex 16.2 System detected with the ABI PRISM 310 Genetic Analyzer. All 16 loci were amplified in a single reaction and detected in a single capillary. The fluorescein-labeled loci (D3S1358, THO1, D21S11, D18S51 and Penta E) are displayed in blue, the TMR labeled loci (Ame-logenin, VWA03, D8S1179, TPOX and FGA) are displayed in black, and the loci labeled with a new dye (D5S818, D13S317, D7S820, D16S539, CSF1PO and Penta D) are displayed in green.The fragments of the prototype ILS-500 size marker are labeled with CRX and are shown in red.(Taken from Promega promotional literature.)
Nature of STR Loci and Nomenclature
The loci DQA1, LDLR, GYPA, HBGG, D7S8 and D1S80 are all, with the exception of D1S80, sequence polymorphisms with defined alleles. The alleles have a simple nomenclature, or naming system. For example LDLR has two alleles, LDLR’A and LDLR’B; these can produce three phenotypes LDLR A, LDLR A,B and LDLR B. In the absence of family studies, we can only determine a phenotype. A phenotype is what is observed. A genotype is inferred from evidence that someone has two copies of an allele, or is homozygous. Thus in a family study it is possible to deduce that someone who is phenotype or type LDLR A is in fact LDLR Al A. When we are not doing family testing we cannot deduce this. Secondly, and perhaps more importantly, it is much easier to identify an apparent homozygote by using the type LDLR A.
The LTR AFLP D1S80 uses a nomenclature based on repeat units. It has been shown that people normally have between 14 and 41 repeats. Irregular repeats have not been found for D1S80. Occasionally, off-ladder electrophoretic variants alleles are encountered (alleles that do not line up with the allele ladder bands), but they usually reflect sequence changes within a repeat affecting electrophoretic properties on acrylamide gels and not variation in repeat size. In general, STR loci behave in the same fashion. However, irregular repeats do occur and the nomenclature has to compensate for that. A common allele at the THO1 locus has 10 repeats. Nine have 4 bp, but one has only 3 bp. Initially this allele was called 10-, which was not consistent with the International System of Genetic Nomenclature. The use of 9.3 indicates that there are nine regular repeats and one with 3 bp. The reporting of STR AFLP types are thus THO1 9, THO1 9,9.3 and THO1 9.3, to be analogous to the LDLR situation. These irregularities can take many different forms. Of the loci included in the CODIS 13, D18S15, D21S11, FGA and THO1,all have irregular repeat alleles. Most of them are .2 alleles, though .1 and .3 variants also occur – some with high frequency, e.g. THOl 9.3.
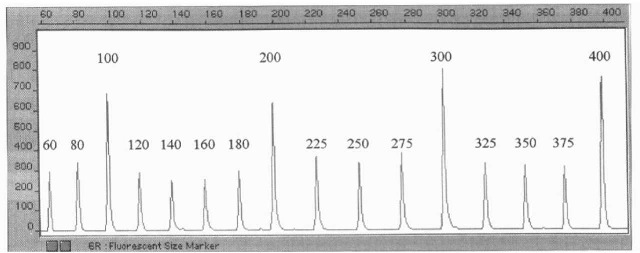
Figure 5 Electropherogram of Promega CRX labeled 60-400 bp ladder as visualized with the PE Biosystems ABI Prism 377 Sequencer.
Usefulness in Detecting Mixtures
One of the major problems in the analysis of forensic evidence is posed by samples containing biological material from more than one source. Polymarker (PM) is very poor at sorting out mixtures because of the limited number of alleles present. DQA1 is slightly better, with its six alleles. D1S80 works well but is limited as a single LTR locus that does not often work well with degraded samples. In contrast, the larger number of discrete alleles at multiple loci make STR multiplexes an excellent tool for identifying components of mixtures. Figure 6 shows the ‘Green’ loci from Profiler Plus from an item of case evidence indicating a mixture. With the quantitative aspects of the electropherogram it is possible to determine the major and minor contributors to the evidence. In the example in Fig. 6, the major contributor is female (X),D8S1179 10,15, D21S11 29,30 and D18S51 14,19,while the minor component is male (X,Y), D8S1179 13,?, D21S11 28,? and D18S51 12,15.
Statistics and Individualization
The calculation of population frequencies, likelihood of a match or probability of source are described elsewhere. The ability to calculate population frequencies for the individual phenotypes observed and the multilocus genotypic arrays all depend on the availability of databases. Often a laboratory will generate and use a database. In the United States the Federal Bureau of Investigation (FBI) recently published a validated database for the 13 CODIS loci. Most laboratories in the United States use this database.
To look at the level of individualization generated by the CODIS 13 loci, the same 19 individuals described in DNA (c) PCR for DQA1, PM, D1S80 and 5 RFLP loci were tested for the CODIS 13 loci ( Table 5). The most common phenotype at each locus was detected for each of the loci. The actual ‘least common phenotype’, created by the co-occurrence of two rare alleles, was not detected for any of the loci, so the least common phenotype presented is that for the 19 samples and not the rarest combination one could find. The most common profile multilocus array represents the worst case scenario and the worst number obtainable if all loci were detected.
Individualization
In the United States the FBI has started releasing reports indicating that biological material originated from a specific source, much as fingerprint examiners have done for many years. The FBI has decided that if the population frequency exceeds 1 in 360 billion the sample is individualized. Other laboratories have chosen high threshold levels such as 1 in 600 billion. Whatever the level chosen, it is apparent that the median multilocus phenotype frequency exceeds both of these thresholds of individualization. The most common multilocus phenotype frequency does not exceed the threshold. None of the 19 individuals was under the individualization threshold of 3.6E+11 (360 billion). In fact the most common profile among the 19 Europeans was 3.113E+13, which is almost 100 times more than the threshold. Thus, although it is possible that individualization may not occur using the 13 CODIS loci, it appears to be relatively unlikely. The frequency of that most common phenotype occurring is the observed frequency, or about one in 200 billion individuals.
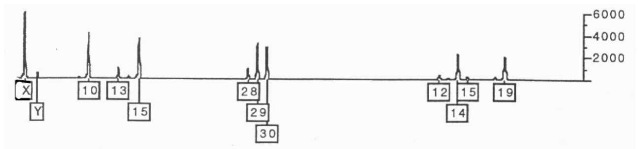
Figure 6 Green loci from a PE Biosystems ABI Prism 310 Genetic Analyzer on an item of evidence containing a mixture.
Table 5 Levels of individualization seen with three different test batteries in 19 European American individuals.
| Test battery | DQA1/PM/D1S80 | 5 RFLP loci | 13 CODIS loci |
| Median (n = 19) | 328000 | 15 900000 | 5.028E+15 |
| MCP | 8900 | 743000 | 1.918E+11 |
| LCP | 73800000 | 1 240000000 | 3.788E+25 |
