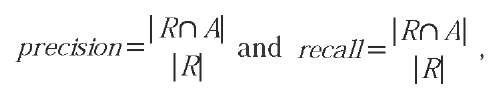Abstract
The topic shows how modern information retrieval methodologies can open up new possibilities to support knowledge management in healthcare. Recent advances in hospital information systems lead to the acquisition of huge quantities of data, often characterized by a high proportion of free narrative text embedded in the electronic health record. We point out how text mining techniques augmented by novel algorithms that combine artificial neural networks for the semantic organization of non-crisp data and hyperbolic geometry for an intuitive navigation in huge data sets can offer efficient tools to make medical knowledge in such data collections more accessible to the medical expert by providing context information and links to knowledge buried in medical literature databases.
introduction
During the last years the development of electronic products has lead to a steadily increasing penetration of computerized devices into our society. Personal computers, mobile computing platforms, and personal digital assistants are becoming omnipresent. With the advance of network aware medical devices and the wireless transmission of health monitoring systems pervasive computing has entered the healthcare domain (Bergeron, 2002).
Consequently, large amounts of electronic data are being gathered and become available online. In order to cope with the sheer quantity, data warehousing systems have been built to store and organize all imaginable kinds of clinical data (Smith & Nelson, 1999).
In addition to numerical or otherwise measurable information, clinical management involves administrative documents, technical documentation regarding diagnosis, surgical procedure, or care. Furthermore, vast amounts of documentation on drugs and their interactions, descriptions of clinical trials or practice guidelines plus a plethora of biomedical research literature are accumulated in various databases (Tange, Hasman, de Vries, Pieter, & Schouten, 1997; Mc Cray & Ide, 2000).
The storage of this exponentially growing amount of data is easy enough: From 1965—when Gordon Moore first formulated his law of exponential growth of transistors per integrated circuit—to today: “Moore’s Law” is still valid. Since his law generalizes well to memory technologies, we up to now have been able to cope with the surge of data in terms of storage capacity quite well. The retrieval of data however, is inherently harder to solve. For structured data such as the computer-based patient record, tools for online analytical processing are becoming available. But when searching for medical information in freely formatted text documents, healthcare professionals easily drown in the wealth of information: When using standard search engine technologies to acquire new knowledge, thousands of “relevant” hits to the query string might turn up, whereas only a few ones are valuable within the individual context of the searcher. Efficient searching of literature is therefore a key skill for the practice of evidence-based medicine (Doig & Simpson, 2003).
Consequently, the main objective of this topic is to discuss recent approaches and current trends of modern information retrieval, how to search very large databases effectively. To this end, we first take a look at the different sources of information a healthcare professional has access to. Since unstructured text documents introduce the most challenges for knowledge acquisition, we will go into more detail on properties of text databases considering MEDLINE as the premier example. We will show how machine learning techniques based on artificial neural networks with their inherent ability for dealing with vague data can be used to create structure on unstructured databases, therefore allowing a more natural way to interact with artificially context-enriched data.
sources of information in healthcare
The advance of affordable mass storage devices encourages the accumulation of a vast amount of healthcare related information. From a technical point of view, medical data can be coarsely divided into structured and unstructured data, which will be illustrated in the following sections.
Data warehouses and clinical Information systems
One major driving force for the development of information processing systems in healthcare is the goal to establish the computer-based patient record (CPR). Associated with the CPR are patient registration information, such as name, gender, age or lab reports such as blood cell counts, just to name a few. This kind of data is commonly stored in relational databases that impose a high degree of structure on the stored information. Therefore, we call such data “structured.” For evidence based medicine the access to clinical information is vital. Therefore, a lot of effort has been put into the goal to establish the computer-based patient record (CPR) as a standard technology in healthcare. In the USA, the Institute of Medicine (1991) defined the CPR as “an electronic patient record that resides in a system specifically designed to support users by providing accessibility to complete and accurate data, alerts, reminders, clinical decision support systems, links to medical knowledge, and other aids.” However, as Sensmeier (2003) states, “this goal remains a vision [...] and the high expectations of these visionaries remain largely unfulfilled.” One of the main reasons why “we are not there yet” is a general lack of integration.
A recent study by Microsoft1 identified as much as 300 isolated data islands within a medical center. The unique requirements of different departments such as laboratory, cardiology, or oncology and the historical context within the hospital often results in the implementation of independent, specialized clinical information systems (CIS) which are difficult to connect (McDonald, 1997). Consequently, main research thrusts are strategies to build data warehouses and clinical data repositories (CDR) from disjoint “islands of information” (Smith & Nelson, 1999). The integration of the separated data sources involves a number of central issues:
1. Clinical departments are often physically separated. A network infrastructure is necessary to connect the disparate sites. Internet and Intranet tools are regarded as standard solutions.
2. Due to the varying needs of different departments, they store their data in various formats such as numerical laboratory values, nominal patient data or higher dimensional CT data. Middleware solutions are required to translate these into a consistent format that permits interchange and sharing.
3. In the real world, many data records are not suitable for further processing. Missing values or inconsistencies have to be treated: Some fields such as blood cell counts are not always available or data was entered by many different persons, and is thus often furnished with a “personal touch.” Cleaning and consistency checks are most commonly handled by rule-based transformations.
4. Departmental workflow should not be strained by data copying tasks. Automated script utilities should take care of the data migration process and copy the data “silently in the background.”
None of the steps above is trivial and certainly it requires a strong management to bring autonomous departments together and to motivate them to invest their working power for a centralized data warehouse. But once such a centralized repository of highly structured data is available it is a very valuable ground for retrospective analysis, aggregation and reporting. Online analytical processing (OLAP) tools can be used to perform customized queries on the data while providing “drill downs” at various levels of aggregation. A study with an interactive visualization framework has shown that it can massively support the identification of cost issues within a clinical environment (Bito, Kero, Matsuo, Shintani, & Silver, 2001). Silver, Sakata, Su, Herman, Dolins, and O’Shea (2001) have demonstrated that data mining “helped to turn data into knowledge.” They applied a patient rule induction method and discovered a small subgroup of inpatients on the DRG level which was responsible for more than 50 percent of revenue loss for that specific DRG (diagnoses related group). The authors describe a “repeatable methodology” which they used to discover their findings: By using statistical tools they identified regions and patterns in the data, which significantly differentiated them from standard cases. In order to establish the reason for increased costs in those regions, they found it necessary that a human expert further analyzes the automatically encircled data patterns. Therefore, a closed loop between clinical data warehouse, OLAP tool and human expert is necessary to gain the maximum of knowledge from the available data.
A recently proposed data mart by Arnrich, Walter, Albert, Ennker, and Ritter (2004) targets in the same direction: Various data sources are integrated into a single data repository. Results from statistical procedures are then displayed in condensed form and can be further transformed into new medical knowledge by a human expert.
unstructured data in biomedical databases
As the discussion on hospital information systems shows, interactive data processing tools form a large potential to extract hidden knowledge from large structured data repositories. A yet more challenging domain for knowledge acquisition tasks is the field of unstructured data. In the following we will briefly present the two premier sources of unstructured data available to the healthcare professional.
Narrative Clinical Data
Many hospitals do not only store numerical patient data such as laboratory test results, but also highly unstructured data such as narrative text, surgical reports, admission notes, or anamneses. Narrative text is able to capture human-interpretable nuances that numbers and codes cannot (Fisk, Mutalik, Levin, Erdos, Taylor, & Nadkarni, 2003). However, due to the absence of a simple machine-parsable structure of free text, it is a challenging task to design information systems allowing the exchange of such kind of information.
In order to address the interoperability within and between healthcare organizations widely accepted standards for Electronic Health Records (EHR) are important. Health Level 7 (HL7)—an ANSI accredited organization, and the European CEN organization both develop standards addressing this problem. The HL7 Clinical Document Architecture (CDA) framework “stems from the desire to unlock a considerable clinical content currently stored in free-text clinical notes and to enable comparison of content from documents created on information systems of widely varying characteristics.” (Dolin, Alschuler, Beebe, Biron, Boyer, Essin, Kimber, Lincoln, & Mattison,, 2001). The emerging new standard based on HL7 version 3 is purely based on XML and therefore greatly enhances the interoperability of clinical information systems by embedding the unstructured narrative text in a highly structured XML framework. Machine-readable semantic content is added by the so-called HL7 Reference Information Model (RIM). The RIM is an all-encompassing look at the entire scope of healthcare containing more than 100 classes with more than 800 attributes. Similar to HL7 v3, the European ENV 13606 standard as defined by the TC 251 group of the CEN addresses document architecture, object models and information messages. For a more detailed discussion on healthcare information standards see Spyrou, Bamidis, Chouvarda, Gogou, Tryfon, and Maglaveras (2002).
Another approach to imprint structure on free text is the application of ontologies to explicitly formalize logical and semantic relationships. The W3C consortium has recently (as of May 2004) recommended the Web Ontology Language (OWL) as a standard to represent semantic content of web information and can therefore be seen as a major step towards the “semantic Web” (Berners-Lee, Hendler, & Lassila, 2001). In the medical domain, the Unified Medical Language System (UMLS) uses a similar technology to represent biomedical knowledge within a semantic network (Kashyap, 2003). Once an ontology for a certain domain is available, it can be used as a template to transfer the therein contained explicit knowledge to corresponding terms of free text from that domain. For example, the term “blepharitis” is defined as “inflammation of the eyelids” in the UMLS. If a medic professional would search for the term “eyelid” within the hospital’s EHRs, documents containing the word “blepharitis” would not show up. However with knowledge from the UMLS, these documents could be marked as relevant to the query. However, the tagging of free text with matching counterparts from an ontology is either a cost intensive human labor or requires advanced natural language processing methods. We shall see below, how computational approaches could address the problem of adding semantics to free narrative text.
Biomedical Publications
For most of the time in human history, the majority of knowledge acquired by mankind was passed on by the means of the written word. It is a natural scientific process to formulate new insights and findings and pass them to the community by publishing. Consequently, scientific literature is constantly expanding. It is not only expanding at growing speed, but also increasing in diversification, resulting in highly specialized domains of expertise.
The MEDLINE database at the U.S. National Library of Medicine (NLM) has become the standard bibliographic database covering the fields of medicine, nursing, veterinary medicine, dentistry, pharmacology, the healthcare system, and the preclinical sciences. The majority of publications in MEDLINE are regular journals and a small number of newspapers and magazines. The database is updated on a regular daily basis from Tuesdays to Saturdays and on an irregular basis in November and December. At the time of this writing, MEDLINE contains approximately 12 million references to articles from over 37,000 international journals dating back to 1966. In 1980 there were about 100,000 articles added to the database, and in 2002 the number increased to over 470,000 newly added entries. That means, currently over 50 new biomedical articles are published on an hourly basis. Clearly, no human is able to digest one scientific article per minute 24 hours a day, seven days a week. But the access to the knowledge within is lively important nonetheless: “The race to a new gene or drug is now increasingly dependent on how quickly a scientist can keep track of the voluminous information online to capture the relevant picture … hidden within the latest research articles” (Ng & Wong, 1999).
Not surprisingly, a whole discipline has emerged which covers the field of “information retrieval” from unstructured document collections, which takes us to the next section.
computational approaches to handle text databases
The area of information retrieval (IR) is as old as man uses libraries to store and retrieve books. Until recently, IR was seen as a narrow discipline mainly for librarians. With the advent of the World Wide Web, the desire to find relevant and useful information grew to a public need. Consequently, the difficulties to localize the desired information “have attracted renewed interest in IR and its techniques as promising solutions. As a result, almost overnight, IR has gained a place with other technologies at the center of the stage” (Baeza-Yates & Ribeiro-Neto, 1999).
Several approaches have shown that advanced information retrieval techniques can be used to significantly alleviate the cumbersome literature research and sometimes even discover previously unknown knowledge (Mack & Hehenberger, 2002). Swanson and Smalheiser (1997) for example designed the ARROWSMITH system which analyses journal titles on a keyword level to detect hidden links between literature from different specialized areas of research. Other approaches utilize MeSH headings (Srinivasan & Rindflesch, 2002) or natural language parsing (Libbus & Rindflesch, 2002) to extract knowledge from literature. Rzhetsky, Iossifov, Koike, Krauthammer, Kra, Morris, Yu, Duboue, Weng, Wilbur, Hatzivassiloglou, and Friedman (2004) have recently proposed an integrated system which visualizes molecular interaction pathways and therefore has the ability to make published knowledge literally visible.
In the following sections we go into more detail and look how computational methods can deal with text.
Representing Text
The most common way to represent text data in numerical form such that it can be further processed by computational means is the so-called “bag of words” or “vector space” model. To this end a standard practice composed of two complementary steps has emerged (Baeza-Yates & Ribeiro-Neto, 1999):
1. Indexing: Each document is represented by a high-dimensional feature vector in which each component corresponds to a term from a dictionary and holds the occurrence count of that term in the document. The dictionary is compiled from all documents in the database and contains all unique words appearing throughout the collection. In order to reduce the dimensionality of the problem, generally only word stems are considered (i.e., the words “computer”, “compute”, and “computing” are reduced to “comput”). Additionally, very frequently appearing words (like “the” & “and”) are regarded as stop words and are removed from the dictionary, because they do not contribute to the distinguishability of the documents.
2. Weighting: Similarities between documents an then be measured by the Euclidean distance or the angle between their corresponding feature vectors. However, this ignores the fact that certain words carry more information content than others. The so-called tf-idf weighting scheme (Salton & Buckley, 1988) has been found a suitable means to deal with this problem: Here the term frequencies (tf) in the document vector are weighted with the inverse document frequency idfi = log(N/sdi), where N is the number of documents in the collection, and sd i denotes the number of times the word stem si occurs in document d.
Although the ” bag of words” model completely discards the order of the words in a document, it performs surprisingly well to capture its content. A major drawback is the very high dimensionality of the feature vectors from which some computational algorithms suffer. Additionally, the distance measure typically does not take polysemy (one word has several meanings) and synonymy (several words have the same meaning) into account.
Evaluating Retrieval Performance
In order to evaluate an IR system, we need some sort of quality measure for its retrieval performance. The most common and intuitively accessible measures are precision and recall. Consider the following situation: We have a collection of C documents and a query Q requesting some information from that set. Let R be the set of relevant documents in the database to that query, and A be the set of documents delivered by the IR system as answers to the given query Q. The precision and recall are then defined by
where RniA is the intersection of the relevant and the answer set, that is, the set of found relevant documents. The ultimate goal is to achieve a precision and recall value of one, that is, to find only the relevant documents, and all of them. A user will generally not inspect the whole answer set A returned by an IR system—who would want to click through all of the thousands of hits typically returned by a web search? Therefore, a much better performance description is given by the so-called precision-recall-curve. Here, the documents in A are sorted according to some ranking criterion and the precision is plotted against the recall values obtained when truncating the sorted answer set after a given rank—which is varied between one and the full size of A. The particular point on the curve where the precision equals the recall is called break even point and is frequently used to characterize an IR system’s performance with a single value.
Therefore, the quality of a search engine which generates hit lists is strongly dependent on its ranking algorithm. A famous example is PageRank, which is part of the algorithm used by Google. In essence, each document (or page) is weighted with the number of hyperlinks it receives from other web pages. At the same time the links themselves are weighed by the importance of the linking page (Brin & Page, 1998). The higher a page is weighted, the higher it gets within the hit list. The analogue to hyperlinks in the scientific literature are citations, that is, the more often an article is cited, the more important it is (Lawrence, Giles & Bollacker, 1999).
The Role of Context
So, we can handle unstructured text and measure how good an IR system works, but a critical question was not answered yet: How do we determine, which documents from a large collection are actually relevant to a given query? To answer this question, we have to address the role of context.
user context
Consider the following example: A user enters the query “Give me all information about jaguars.” The set of relevant documents will be heavily dependent on the context in which the user phrases this query: A motor sports enthusiast is probably interested in news about the “Jaguar Racing” Formula 1 racing stable, a biologist probably wants to know more about the animal. So, the context—in the sense of the situation in which the user is immersed—plays a major role (Johnson, 2003; Lawrence, 2000).
Budzik, Hammond and Birnbaum (2001) discuss two common approaches to handle different user contexts:
1. Relevance feedback: The user begins with a query and then evaluates the answer set. By providing positive or negative feedback to the IR system, this can modify the original query by adding positive or negative search terms to it. In an iterative dialogue with the IR system, the answer set is then gradually narrowed down to the relevant result set. However, as studies (Hearst, 1999) have shown, users are generally reluctant to give exhaustive feedback to the system.
2. Building user profiles: Similar to the relevance feedback, the IR system builds up a user profile across multiple retrieval sessions, that is, with each document the user selects for viewing, the profile is adapted. Unfortunately, such a system does not take account of “false positives”, that is, when a user follows a link that turned out to be of no value when inspecting it closer. Additionally, such systems integrate short term user interests into accumulated context profiles, and tend to inhibit highly specialized queries which the user is currently interested in.
Budzik, Hammond & Birnbaum (2001) presented a system that tries to guess the user context from open documents currently edited or browsed on the work space. Their system constitutes an “Information Management Assistant [which] observes user interactions with everyday applications and anticipates information needs [.] in the context of the user’s task.” Another system developed by Finkelstein and Gabrilovich (2002) analyzes the context in the immediate vicinity of a user-selected text, therefore making the context more focused. In an evaluation of their system both achieved consistently better results than standard search engines are able to achieve without context.
Document context
Context can also be seen from another point of view: Instead of worrying about the user’s intent and the context in which a query is embedded, an information retrieval system could make the context in which retrieved documents are embedded more explicit. If each document’s context is immediately visible to the information seeker, the relevant context might be quickly picked out. Additionally, the user might discover a context he had not in mind when formulating the query and thus find links between his intended and an unanticipated context. In the following we describe several approaches that aim in this direction.
Augmenting Document sets with context
Text Categorization
One way to add context to a document is by assigning a meaningful label to it (Le & Thoma, 2003). This constitutes a task of text categorization and there exist numerous algorithms that can be applied. The general approach is to select a training set of documents that are already labelled. Based on the “bag of words” representation, machine learning methods learn the association of category labels to documents. For an in depth review of statistical approaches (such as naives Bayes or decision trees) see Yang (1999). Computationally more advanced methods utilize artificial neural network architectures such as the support vector machine which have achieved break even values close to 0.9 for the labelling of news wire articles (Joachims, 1998; Lodhi, 2001).
However, in the medical domain, 100 categories are seldom adequate to describe the context of a text. In case of the MEDLINE database, the National Library of Medicine has developed a highly standardized vocabulary, the Medical Subject Headings (MeSH) (Lowe & Barnett, 1994). They consist of more than 35,000 categories that are hierarchically organized and constitute the basis for searching the database. To guarantee satisfactory search results of constant quality, reproducible labels are an important prerequisite. However, the cost of human indexing of the bio-medical literature is high: according to Humphrey (1992) it takes one year to train an expert the task of document labelling. Additionally, the labelling process lacks a high degree of reproducibility. Funk, Reid, and McGoogan (1983) have reported a mean agreement in index terms ranging from 74 percent down to as low as 33 percent for different experts. Because the improvement of index consistency is such demanding, assistance systems are considered to be a substantial benefit. Recently, Aronson, Bodenreider, Chang, Humphrey, Mork, Nelson, Rindflesh, and Wilbur (2000) have presented a highly tuned and sophisticated system which yields very promising results. Additionally to the bag of words model their system utilizes a semantic network describing a rich ontology of biomedical knowledge (Kashyap, 2003).
Unfortunately, the high complexity of the MeSH terms makes it hard to incorporate a MeSH-based categorization into a user interface. When navigating the results of a hierarchically ordered answer set it can be a time-consuming and frustrating process: Items which are hidden deep within the hierarchy can often only be obtained by descending a tree with numerous mouse-clicks. Selection of a wrong branch requires backing up and trying a neighboring branch. Since screen space is a limited resource only a small area of context is visible and requires internal “recali-bration” each time a new branch is selected. To overcome this problem, focus and context techniques are considered to be of high value. These are discussed in more detail below.
The Cluster Hypothesis
An often cited statement is the cluster hypothesis, which states that documents which are similar in their bag of words feature space tend to be relevant to the same request (van Rijsbergen, 1979). Leuski (2001) has conducted an experiment where an agglomerative clustering method was used to group the documents returned by a search engine query. He presented the user not with a ranked list of retrieved documents, but with a list of clusters, where each cluster in turn was arranged as a list of documents. The experiment showed, that this procedure “can be much more helpful in locating the relevant information than the traditional ranked list.” He could even show that the clustering can be as effective as the relevance feedback methods based on query expansion. Other experiments also validated the cluster hypothesis on several occasions (Hearst & Pedersen, 1996; Zamir & Etzioni, 1999). Recently the vivisimo1 search engine has drawn attention by utilizing an online clustering of retrieval sets, which also includes an interface to PubMed / MEDLINE.
Visualizing context
Another way to create context in line with the spirit of the cluster hypothesis is by embedding the document space in a visual display. By making the relationship between documents visually more explicit, such that the user can actually see inter-document similarities, the user gets (i) an overview of the whole collection, and (ii) once a relevant document has been found, it is easier to locate others, as these tend to be grouped within the surrounding context of already identified valuable items. Fabrikant and Buttenfield (2001) provide a more theoretical framework for the concept of “spatialization” with relation to cognitive aspects and knowledge acquisitions: Research on the cognition of geographic information has been identified as being important in decision making, planning and other areas involving human-related activities in space.
In order to make use of “spatialization”, that is, to use cognitive concepts such as “nearness” we need to apply some sort of transformation to project the documents from their high-dimensional (typically several thousands) bag-of-words space onto a two dimensional canvas suitable for familiar inspection and interaction. The class of algorithms performing such a projection is called multi dimensional scaling (MDS). A large number of MDS methods such as Sammon mapping, spring models, projection pursuit or local linear embeddings have been proposed over the years. Skuping and Fabrikant (2003) provide an excellent discussion of the most common MDS variants in relation to spatialized information visualization.
Self-Organizing Maps
The notion of the self-organizing map (SOM) has been introduced by Kohonen (1982) more than 20 years ago. Since then is has become a well-accepted tool for exploratory data analysis and classification. While applications ofthe SOM are extremely wide spread—ranging from medical imaging, classification of power consumption profiles, or bank fraud detection—the majority of uses still follows its original motivation: to use a deformable template to translate data similarities into spatial relations.
The following example uses a simple toy dataset to demonstrate the properties of the SOM algorithm. Consider a 13-dimensional dataset describing animals with properties such as small, medium, big, two legs, hair, hooves, can fly, can swim, and so on. When displaying the data as a large table, it is quite cumbersome to see the interrelationship between the items, that is, animals. The map shown in Figure 1 depicts a trained SOM with neurons placed on a 20×20 regular grid. After the training process, each animal is “presented” to the map, and the neuron with the highest activity gets labelled with the corresponding name. As can be seen in the figure the SOM achieves a semantically reasonable mapping of the data to a two-dimensional “landscape”: birds and non-birds are well separated and animals with identical features get mapped to identical nodes.
Figure 1. A map of animals
There have been several approaches to use the SOM to visualize large text databases and relations of documents therein. The most prominent example is the WEBSOM project. Kohonen, Kaski and Lagus, (2000) performed the mapping of 7 million patent abstracts and obtained a semantic landscape of the corresponding patent information. However, screen size is a very limited resource, and as the example with 400 neurons above suggests, a network with more than one million nodes—2500 times that size—becomes hard to visualize: When displaying the map as a whole, annotations will not be readable, and when displaying a legible subset, important surrounding context will be lost. Wise (1999) has impressively demonstrated the usefulness of compressed, map-like representations of large text collections: His ThemeView reflects major topics in a given area, and a zoom function provides a means to magnify selected portions of the map—unfortunately without a coarser view to the surrounding context.
Focus & Context Techniques
The limiting factor is the two dimensional Euclidean space we use as a data display: The neighborhood that “fits” around a point is rather restricted: namely by the square of the distance to that point. An interesting loophole is offered by hyperbolic space: it is characterized by uniform negative curvature and results in a geometry, where the neighborhood around a point increases exponentiallyw ith the distance. This exponential behavior was firstly exploited (and patented) by the “hyperbolic tree browser” from Lamandpng & Rao (1994), followed by a Web content viewer by Munzner (1998). Studies by Pirolli, Card and van der Wege (2001) showed that the particular focus & context property offered by hyperbolic space can significantly accelerate “information foraging”.
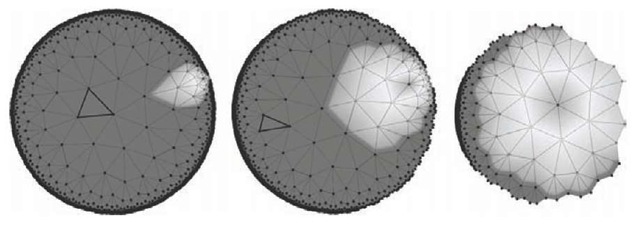
Naturally, it becomes apparent to combine the SOM algorithm with the favorable properties of hyperbolic space (Ritter, 1999). The core idea of the hyperbolic self-organizing map (HSOM) is to employ a grid of nodes in the hyperbolic plane H2. For H2 there exists an infinite number of tessellations with congruent polygons such that each grid point is surrounded by the same number of neighbors (Magnus, 1974). As stated above, an intuitive navigation and interaction methodology is a crucial element for a well-fitted visualization framework. By utilizing the Poincare projection and the set of Mobius transformations (Coxeter, 1957) a “fish-eye” fovea can be positioned on the HSOM grid allowing an intuitive interaction methodology. Nodes within the fovea are displayed with high resolution, whereas the surrounding context is still visible in a coarser view. For further technical details of the HSOM see (Ritter, 1999; Ontrup & Ritter, 2001). An example for the application of the fish-eye view is given in Figure 2. It shows a navigation sequence where the focus was moved towards the highlighted region of interest. Note, that from the left to the right details in the target area get increasingly magnified, as the highlighted region occupies more and more display space. In contrast to standard zoom operations, the current surrounding context is not clipped, but remains gradually compressed at the periphery of the field of view. Since all operations are continuous, the focus can be positioned in a smooth and natural way.
Figure 2. Navigation snapshot showing isometric transformation of HSOM tessellation. The three images were acquired while moving the focus from the center of the map to the highlighted region at the outer perimeter. Note the “fish-eye” effect: All triangles are congruent, but appear smaller as further they are away from the focus.
Browsing Medline in Hyperbolic Space
The following example presents a HSOM framework that combines the aspects of information retrieval, context enrichment and intuitive navigation into a single application. The following images show a two dimensional HSOM, where the nodes are arranged on a regular grid consisting of triangles similar to those in Figure 2. The HSOM’s neurons can be regarded as “containers” holding those documents for which the corresponding neuron is “firing” with the highest rate. These containers are visualized as cylinders embedded in the hyperbolic plane. We can then use visual attributes of these containers and the plane to reflect document properties and make context information explicit:
• The container sizes reflect the number of documents situated in the corresponding neuron.
• A color scale reflects the number of documents which are labelled with terms from the Anatomy hierarchy of MeSH terms, i.e. the brighter a container is rendered, the more articles labelled with Anatomy MeSH entries reside within that node.
• The color of the ground plane reflects the average distance to neighboring nodes in the semantic bag-of-words space. This allows the identification of thematic clusters within the map.
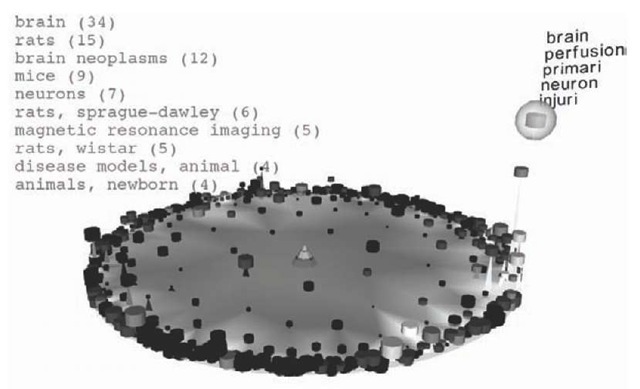
Figure 3 shows a HSOM that was trained with approximately 25,000 abstracts from MEDLINE. The interface is split into two components: the graphical map depicted below and a user interface for formulating queries (not shown here). In the image below, the user has entered the search string “brain”. Subsequent to the query submission, the system highlights all nodes belonging to abstracts containing the word “brain” by elevating their corresponding nodes. Additionally the HSOM prototype vectors—which were autonomously organized by the system during a training phase—are used to generate a key word list which annotate and semantically describe the node with the highest hit rate. To this end, the words corresponding to the five largest components of the reference vector are selected. In our example these are: brain, perfusion, primari, neuron, and injuri. In the top left the most prominent MeSH terms are displayed, that is, 34 articles are tagged with the medical subject heading “brain”, 15 are tagged with “rats”, and so on.
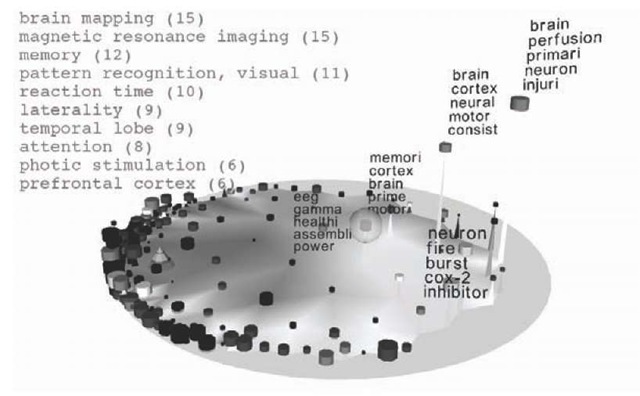
The map immediately shows that there are several clusters of brain-related articles in the right part of the map. Moving the focus into this area leads to the situation shown in Figure 4. By selecting single neurons with the mouse, the user can selectively retrieve key words of individual containers. Additionally, the MeSH terms of the selected node are displayed. As can be seen from the keyword distribution, all nodes share semantically related characteristics. The two leftmost labelled nodes do not contain key words like “brain” or “neuron”, but the labelling with “cord”, “spinal”, “eeg”, and “gamma”, respectively, indicates a close relation to the selected “brain-area”.
Figure 3. MEDLINE articles mapped with the HSOM. The user submitted the query string “brain” and the corresponding results are highlighted by the system.
The drill-down process to the document level combines the strengths of the graphical and the textual display: After zooming-in on a promising region, the titles of documents pertaining to a particular node can be displayed by a double mouse click. Note, that due to the ordering process of the SOM algorithm these documents are typically very similar to each other—and in the sense of the clustering hypothesis therefore might all provide an answer to the same query. A further double click on a document title then displays the selected abstract on the screen. In this way, the hyperbolic self-organizing map can provide a seamless interface offering rapid overview of large document collections while allowing at the same time the drill down to single texts.
benefits for a hospital information system
In the framework of a hospital information system, such a context enriched interactive document retrieval system might be of great benefit to the clinical personal. According to Chu and Cesnik (2001) a hospital may generate up to five terabytes of data a year, where 20 to 30 percent of such data are stored as free text reports such as medical history, assessment and progress notes, surgical reports or discharge letters. Free text fields in medical records are considered of invaluable importance for medical relevance (Stein, Nadkarni, Erdos, & Miller, 2000). By using document standards like the CDA as briefly discussed in Section 2.2, the clinician is able to browse and retrieve the data according to the structure provided by the document framework. However, many existing information systems do not yet take advantage of the complex CDA or the even more substantial Reference Information Model—because it is a very time consuming task to transfer documents to the new standards and to fit the enormous mass of model objects to free text data.
Nevertheless, these systems store valuable information—just in a format which cannot be directly analyzed by structured methods. In order to provide access by content, the aforementioned Information Retrieval methodologies can significantly contribute to facilitate these tasks: a self-organizing map creates structure and context purely on the statistical word distributions of free text and thus allows a semantic browsing of clinical documents. This in turn offers the possibility to gain a problem orientated perspective on health records stored in information systems, that is, as Lovis et al (2000) have noted: “intelligent browsing of documents, together with natural language emerging techniques, are regarded as key points, as it appears to be the only pertinent way to link internal knowledge of the patient to general knowledge in medicine.”
Figure 4. The user has moved the focus into the target area and can now inspect the context enriched data more closely.
conclusion
Recent advances in hospital information systems have shown that the creation of organization-wide clinical data repositories (CDR) plays a key role for the support of knowledge acquisition from raw data. Currently, many case studies are published which emphasize that data mining techniques provide efficient means to detect significant pat terns in medical records. Interactive OLAP tools support best practice and allow for informed real time decision-making and therefore build the solid foundation for further improvements in patient care.
The next step to further broaden the knowledge base in healthcare environments will be the integration of unstructured data, predominantly free narrative text documents. The nature of this type of data will demand for new approaches to deal with the inherently vague information contained in these documents. We have shown how artificial neural networks with their ability for self-organization of non-crisp data provide an almost natural link between the hard computational world and the soft- computing of the human brain. We believe that interactive information retrieval methodologies that account for the rich context of natural language will significantly contribute to knowledge acquisition through text analysis.