Introduction
Given the exponential growth rate of medical data and the accompanying biomedical literature, more than 10,000 documents per week (Leroy et al., 2003), it has become increasingly necessary to apply data mining techniques to medical digital libraries in order to assess a more complete view of genes, their biological functions and diseases. Data mining techniques, as applied to digital libraries, are also known as text mining.
Background
Text mining is the process of analyzing unstructured text in order to discover information and knowledge that are typically difficult to retrieve. In general, text mining involves three broad areas: Information Retrieval (IR), Natural Language Processing (NLP) and Information Extraction (IE). Each of these areas are defined as follows:
• Natural Language Processing: a discipline that deals with various aspects of automatically processing written and spoken language.
• Information Retrieval: a discipline that deals with finding documents that meet a set of specific requirements.
• Information Extraction: a sub-field of NLP that addresses finding specific entities and facts in unstructured text.
MAIN THRUST
The current state of text mining in digital libraries is provided in order to facilitate continued research, which subsequently can be used to develop large-scale text mining systems. Specifically, an overview of the process, recent research efforts and practical uses of mining digital libraries, future trends and conclusions are presented.
Text mining process
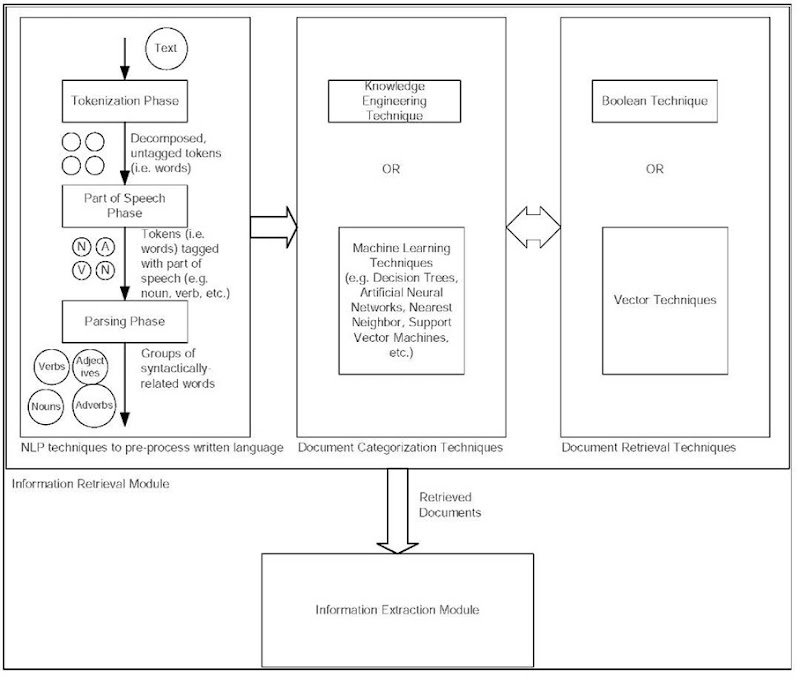
Text mining can be viewed as a modular process that involves two modules: an information retrieval module and an information extraction module. presents the relationship between the modules and the relationships between the phases within the information retrieval module. The former module involves using NLP techniques to pre-process the written language and using techniques for document categorization in order to find relevant documents. The latter module involves finding specific and relevant facts within text. NLP consists of three distinct phases: (1) tokenization, (2) parts of speech (PoS) tagging and (3) parsing. In the tokenization step, the text is decomposed into its subparts, which are subsequently tagged during the second phase with the part of speech that each token represents (e.g., noun, verb, adjective, etc.). It should be noted that generating the rules for PoS tagging is a very manual and labor-intensive task. Typically, the parsing phase utilizes shallow parsing in order to group syntactically related words together because full parsing is both less efficient (i.e., very slow) and less accurate (Shatkay & Feldman, 2003). Once the documents have been pre-processed, then they can be categorized.
There are two approaches to document categorization: Knowledge Engineering (KE) and Machine Learning (ML). Knowledge Engineering requires the user to manually define rules, which can consequently be used to categorize documents into specific pre-defined categories. Clearly, one of the drawbacks of KE is the time that it would take a person (or group of people) to manually construct and maintain the rules. ML, on the other hand, uses a set of training documents to learn the rules for classifying documents.
Figure 1. Overview of text mining process
Specific ML techniques that have successfully been used to categorize text documents include, but are not limited to, Decision Trees, Artificial Neural Networks, Nearest Neighbor and Support Vector Machines (SVM) (Stapley et al., 2002). Once the documents have been categorized, then documents that satisfy specific search criteria can be retrieved.
There are several techniques for retrieving documents that satisfy specific search criteria. The Boolean approach returns documents that contain the terms (or phrases) contained in the search criteria; whereas, the vector approach returns documents based upon the term frequency-inverse document frequency (TF x IDF) for the term vectors that represent the documents. Variations of clustering and clustering ensemble algorithms (Iliopoulous et al., 2001; Hu, 2004), classification algorithms (Marcotte et al., 2001) and co-occurrence vectors (Stephens et al., 2001) have been successfully used to retrieve related documents. An important point to mention is that the terms that are used to represent the search criteria as well as the terms used to represent the documents are critical to successfully and accurately returning related documents. However, terms often have multiple meanings (i.e., polysemy) and multiple terms can have the same meaning (i.e., synonyms). This represents one of the current issues in text mining, which will be discussed in the next section.
The last part of the text mining process is information extraction, of which the most popular technique is co-occurrence (Blaschke & Valencia, 2002; Jenssen et al., 2001). There are two disadvantages to this approach, each of which creates opportunities for further research. First, this approach depends upon assumptions regarding sentence structure, entity names, and etcetera that do not always hold true (Pearson, 2001). Furthermore, this approach relies heavily on completeness of the list of gene names and synonyms and summarizes the modular process of text mining.
Research to Address Issues in mining digital libraries
The issues in mining digital libraries, specifically medical digital libraries, include scalability, ambiguous English and biomedical terms, non-standard terms and structure and inconsistencies between medical repositories (Shatkay & Feldman, 2003). Most of the current text mining research focuses on automating information extraction (Shatkay & Feldman, 2003). The scalability of the text mining approaches is of concern because of the rapid rate of growth of the literature. As such, while most of the existing methods have been applied to relatively small sample sets, there has been an increase in the number of studies that have been focused on scaling techniques to apply to large collections (Pustejovsky et al., 2002; Jenssen et al., 2002). One exception to this is the study by Jenssen et al. (2001) in which the authors used a predefined list of genes to retrieve all related abstracts from PubMed that contained the genes on the predefined list.
Table 1. General text mining process
|
General Step |
General Purpose |
General Issues |
General Solutions |
|
IR |
Identify and retrieve relevant docs |
aliases, synonyms & homonyms |
|
|
IE |
Find specific and relevant facts within text [e.g., find specific gene entities (entity extraction) & relationships between specific genes (relationship extraction)] |
aliases, synonyms & homonyms |
Controlled vocabularies (ontologies) [e.g., gene terms and synonyms: LocusLink (Pruitt & Maglott, 2001), SwissProt (Boeckmann et al., 2003), HUGO (HUGO, 2003), National Library of Medicine's MeSH (NLM, 2003)] |
Since mining digital libraries relies heavily on the ability to accurately identify terms, the issues of ambiguous terms, special jargon and the lack of naming conventions are not trivial. This is particularly true in the case of digital libraries where the issue is further compounded by non-standard terms. In fact, a lot of effort has been dedicated to building ontologies to be used in conjunction with text mining techniques (Boeckmann et al., 2003; HUGO, 2003; Liu et al., 2001; NLM, 2003; Oliver et al., 2002; Pruitt & Maglott, 2001; Pustejovsky et al., 2002). Manually building and maintaining ontologies, however, is a time consuming effort. In light of that, there have been several efforts to find ways of automatically extracting terms to incorporate into and build ontologies (Nenadic et al., 2002; Ono et al., 2001). The ontologies are subsequently used to match terms. For instance, Nenadic et al. (2002) developed the Tagged Information Management System (TIMS), which is an XML-based Knowledge Acquisition system that uses ontology for information extraction over large collections.
Uses of Text Mining in Medical Digital Libraries
There are many uses for mining medical digital libraries that range from generating hypotheses (Srinivasan, 2004) to discovering protein associations (Fu et al., 2003). For instance, Srinivasan (2004) developed MeSH-based text mining methods that generate hypotheses by identifying potentially interesting terms related to specific input. Further examples include, but are not limited to: uncovering uses for thalidomide (Weeber et al., 2003), discovering functional connections between genes (Chaussabel & Sher, 2002) and identifying viruses that could be used as biological weapons (Swanson et al., 2001). He summarizes some of the recent uses of text mining in medical digital libraries.
FUTURE TRENDS
The large volume of genomic data resulting and the accompanying literature from the Human Ge-nomic project is expected to continue to grow. As such, there will be a continued need for research to develop scalable and effective data mining techniques that can be used to analyze the growing wealth of biomedical data. Additionally, given the importance of gene names in the context of mining biomedical literature and the fact that there are a number of medical sources that use different naming conventions and structures, research to further develop ontology will play an important part in mining medical digital libraries. Finally, it is worth mentioning that there has been some effort to link the unstructured text documents within medical digital libraries with their related structured data in data repositories.
Table 2. Some uses of text mining medical digital libraries
|
Application |
Technique |
Supporting Literature |
|
Building gene networks |
Co-occurrence |
Jenssen et al., 2001; Stapley & Be-noit, 2000; Adamic et al., 2002 |
|
Discovering protein association |
Unsupervised cluster learning and vector classification |
Fu et al., 2003 |
|
Discovering gene interactions |
Co-occurrence |
Stephens et al., 2001; Chaussabel & Sher, 2002 |
|
Discovering uses for thalidomide |
Mapping phrases to UMLS concepts |
Weeber et al., 2001 |
|
Extracting and combining relations |
Rule-based parser and co-occurrence |
Leroy et al., 2003 |
|
Generating hypotheses |
Incorporating ontologies (e.g., mapping terms to MeSH) |
Srinivasan, 2004; Weeber et al., 2003 |
|
Identifying biological virus weapons |
|
Swanson et al., 2001 |
conclusion
Given the practical applications of mining digital libraries and the continued growth of available data, mining digital libraries will continue to be an important area that will help researchers and practitioners gain invaluable and undiscovered insights into genes, their relationships, biological functions, diseases and possible therapeutic treatments.
KEY TERMS
Bibliomining: Data mining applied to digital libraries to discover patterns in large collections.
Bioinformatics: Data mining applied to medical digital libraries.
Clustering: An algorithm that takes a dataset and groups the objects such that objects within the same cluster have a high similarity to each other, but are dissimilar to objects in other clusters.
Information Extraction: A sub-field of NLP that addresses finding specific entities and facts in unstructured text.
Information Retrieval: A discipline that deals with finding documents that meet a set of specific requirements.
Machine Learning: Artificial intelligence methods that use a dataset to allow the computer to learn models that fit the data.
Natural Language Processing: A discipline that deals with various aspects of automatically processing written and spoken language.
Supervised Learning: A machine learning technique that requires a set of training data, which consists of known inputs and a priori desired outputs (e.g., classification labels) that can subsequently be used for either prediction or classification tasks.
Unsupervised Learning: A machine learning technique, which is used to create a model based upon a dataset; however, unlike supervised learning, the desired output is not known a priori.