Introduction
The geographic distribution and genetic diversity of dengue virus is deeply rooted in Asia suggesting its origin from this region, with first reported out-break of DHF from Philippine in 1953 (Halstead, 1980). One of the characteristics notable in Asian regions, where the disease is endemic is that dengue hemorrhagic fever outbreaks occur in repetitive cycles of 3-5 years, (Ferguson et al, 1999). The incidence of disease and its severity varies across different dengue virus serotypes and also between primary and secondary infections of same serotypes(Vaughn DW et al).
Due to lack of in-vivo study models, there is little information about factors contributing to disease severity and its variation across dengue virus genotypes and the cyclical nature of dengue outbreaks. It is however critical to study these factors particularly in the South East Asian region where incidence of dengue cases is thought to be associated with variables such as water, sanitation, population density and rate of literacy as opposed to developed countries where ambient temperature, moisture and rainfall perhaps plays the major role. A better understanding of disease epidemiology and pathogenesis will help identify optimum control measures in the region. It will also develop systems for predicting the outcome of mass vaccination when the vaccine becomes available in this region.
The topic has been divided in three parts: the first part will discuss the historical evolution of the dengue virus in the region its spatial and temporal distribution. It will also look at the effects of covariates such as poverty, water supply, sanitation and global warming on expansion of the dengue endemic regions. .
The second part of the topic will focus on the genetic evolution of the viral isolates circulating in the region. Phylogenetic studies of dengue viruses have uncovered genetic variation within each serotypes, these variations have been organized in discrete clusters on dendograms. Analyses of such studies have broadened our horizon to relate the mutational changes with disease evolution and factors like seasonality and incidence variability. This part of topic will focus on the common mutational variations that have been reported so far and how these relate with the disease dynamics in the endemic region. In the third and final part of the topic an attempt has been made to relate the mutational changes of dengue genotypes with disease severity. Vast array of literature has been published investigating relationship of genetic variation with disease severity. The structure of virus E- protein that confers the viral infectivity and host immune response of the virus (E.Descloux,2009) remains the focus of such studies. Sequence variation at different loci such as CprM, E/NS1, preM/E, C/prM/M and untranslated regions etc. have been investigated for its association with disease severity. This part of topic will throw some light on our current understanding of disease severity and it relation with genetic variation.
Historical background of dengue virus in South East Asian
Geographically South East Asia comprises of land south of China to east of India extending as far as to the north of Australia. Although geographically the region is well defined, the list of countries included in this region varies due to political reasons. For the purpose of this review W.H.O based definition has been used. In addition status of dengue virus in further south of the region; including countries like Pakistan and Bangladesh have also been included to encompass the broader spectrum of the region.
Dengue vector evolution
Evidences suggest that vectors Ades aegypti and Ades albopictus originated from darker sylvan forms found in African tropical forests. It is believed to have reached New World from West Africa via slave ships during the 17th century (Gubler, D.J.1998). Adesaegypti was introduced into the coastal cities of South East Asia from East Africa around nineteenth century via the shipping industry. With the eruption of World War II it became deeply entrenched in many cities (Gubler, D.J.1998). The mitochondrial genetic diversity studies have revealed circulation of two distinct clusters of Ades aegypti in South East Asia one with strains from French Polynesia, Guinea and Brazil while the other cluster is of strains that migrated from Europa Island in Mozambique and Amazonia (Mousson et al 2005). In contrast; A. albopictus is known to be native to South East Asia. It has spread within past few decades to various countries primarily due to introduction of trade of used tyres worldwide. Using ecological niche modeling Benedict and co-workers have predicted the risk of global invasion by Ades albopitus secondary to cargo trade and increasing air travel. Although temperate and humid climates are prerequisites for the optimum survival of both the vectors but A. albopictus is known to better acclimatize to the cold and dry weather due to its ability of efficient egg diapause during the extreme conditions, thus favoring its survival in the regions with exotic temperature ranges (Benedict, M.Q, et al 2007).
Factors leading to disease spread in SEA
The factors responsible for the insurmountable expansions of dengue in the region are complex and thought to be intricately linked with vector-host-virus triad, socioeconomic stresses and climatic variations. There are excellent reviews that discuss the impact of these factors in details (Aiken, S.R. 1978, Kendall, C. et al 1991, Halstead, S.B. 1966). Only salient factors in context of SEA will be discussed here. The distribution of DHF outbreaks in SEA correlates with emergence of mosquito A. aegypti in South East Asian countries perhaps due to displacement of indigenous A. albpictus in the region. This is considered to be associated with uncontrolled urbanization leading to shanty towns with inadequate pipe water supply and poor sanitation.
A. albopictus is semi domestic species that breeds on natural and man-made breeding sights; it feeds on variety of animals, birds and man. The A. aegypti on the other hand is more acclimatized to urban set-up, once established the density of this mosquito is directly proportional to density of human population and artificial breeding sites (Merril S.A et al 2005), it feeds almost exclusively on humans. Moreover A. aegypti is considered to be more competent vector for dengue virus. Genetic traits that determines successful midgut infection by DEN virus have been mapped on several loci on A. aegypti chromosomes (Benedict, M.Q, et al 2007) indicating that vector competence is genetically determined. The extent to which these mosquitoes compete with each other in the environment is not clear, nonetheless the balance of two species in the region is important, and the socioeconomic factors in SEA appear to be displacing A.albopictus in favour of A.aegypti leaving the population more susceptible. The poor socioeconomic conditions are major contributing factor to sustained vector activity with severe form of disease in the South East Asia. The breeding habitats of A.aegypti have been strongly associated with squatter settlements, inadequate piped water supply and sewage facilities (Halstead, S.B. 1966). In addition, there are impacts of higher environmental temperature in the region. High temperature is inversely related to the mosquito gonotropic cycle and viral extrinsic incubation period; this increases the egg laying episodes resulting in more blood meals and increased risk for viral transmission. In addition shorten extrinsic viral incubation period culminate to increase virus load at time of inoculation (Focks D.A. et al 1993). These effects have been proven for dengue vectors in simulation studies conducted by (Cox J et al 2001) and it has been projected that increase in global temperature would increase the length of transmission season in temperate regions.
Dengue fever and dengue hemorrhagic fever
The word dengue is believed to have originated from Swahili language "ki denga pepo", which describes sudden cramp like seizure. The clinical symptoms suggestive of dengue virus infection can be traced back to Chinese Chin Dynasty (265-420 AD) where disease was considered as water poison and was known to be associated with water and insects (anonymous 2006).
Emergence of the disease in the new world can be traced back to the transmigration of the vector in the 17th century. There are reports that suggest possible epidemics of dengue like illness in three major continents (Asia, Africa and North America) as early as 1779 and 1780, within Asia Batavia (now known as Jakarta) was affected by this outbreak (Halstead,S.B. 1966). By early nineteenth century Dengue fever was known to be endemic in the rural areas of South East Asia probably due to the indigenous vector A.albopictus. It manifested as self limiting disease to which native population developed immunity at early age. With the advent of A. aegypti at Asian ports, the disease spread to the main inland cities and towns. It is assumed that unlike rural population, the urban populations of South East Asia remained susceptible to dengue virus and were then infected by newly imported vector. Dengue epidemics progressively became less frequent as urban population became immune to the disease, until 1953 when a new form of dengue fever was reported from Thailand and Manila, where children suffered from fever followed by bleeding diathesis; the disease was then called as Philippine Fever (Aiken, S.R. 1978). By 1960′s the hemorrhagic form of disease had spread to Malaysia, Vietnam, Sri Lanka, Singapore and Indonesia (Halstead, S.B. 1966). The disease epidemiology extended and outbreaks of dengue hemorrhagic fever (DHF) were reported from India 1988) French Polynesia (1990), Pakistan (1992) and Bangladesh (2000).Until recently, DHF was considered to be disease of childhood, especially in South
East Asia where mean age of cases under fifteen, and the modal age of five or slightly higher was reported from countries such as Thailand, Philippines and Malaysia, however, recent reports are now documenting increasing number of DHF and DSS in adult population as well (Khan E et al 2007). The precise cause of DHF/DSS remains elusive despite enormous research in this area. Evidences suggest interplay of multiple factors such as host genetic make-up with unique immune response and viral virulence may play a role in determining the severity of the disease.
Pathogenesis of severe dengue disease
There are two form of Severe disease, namely dengue shock syndrome (DSS) and DHF without shock. It is proposed that devastating coagulation derangements due to host immune response leads to heamorrhage and shock in severe cases. The concept of original antibody sin leading to immune enhancement is considered to be the main reason whereby infection with one type of dengue virus sensitizes an individual and that subsequent infection with different virus type elicits a hypersensitivity reaction (secondary infection). Various studies have been conducted to show the association of elevated cytokines in patients presenting with DHF and DSS. Elevated serum levels of cytokine and chemokines such as IL-2, IL-8, IL-6, IL-10, IL-13, TNF and INF-γ have been found to be significantly associated with patients presenting with DHF and DSS in clinical setting (Azeredo et al., 2001; Hung, et al., 2004, Clyde. K. et al., 2006). It has been proposed that the pro-inflammatory cytokines released by the cross reactive memory T-cellls, induce plasma leakage by its effects on the endothelial cells (Eva.H. et al 2004; Aviruntanan et al., 1998). In fact in-vitro studies have rendered endothelial cell monolayers permeable by the application of chemokine such as IL-1ß (Cardier et al.,2005). In vitro-and in-vivo models of studies also suggest role of decreased nitric oxide levels and its relation with IL-10 and raised viral load (Simmons et al., 2007). There is evidence that suggests relation of increased expression of certain cytokines such as IL-1ß, TNF-γ, and IL-6 with elevated NO production (Guzik et al., 2003).
With the advances in genomic and bioinformatics tools the scope of genetic studies has greatly expanded particularly in depth data on genomic changes and its association with disease epidemiology, seasonality and severity has been made available. Growing availability of comparative genome sequence data has provided important insights into the molecular evolution of dengue virus. Evidence strongly suggests appearance of new strains correlating with DHF/DSS epidemics. Despite the wealth of genomic data now available the exact cause and effect of viral virulence and clade changes is yet to be proven, however it is quite evident that different serotypes and viral linage is continually changing with local extinction and emergence of new clade and that the introduction of new clade in the region translates in form of outbreaks of DHF and DSS.
Distribution of dengue virus serotypes in SEA
Dengue like other RNA viruses is prone to genetic mutations as it replicates using RNA-Polymerase; enzyme that lacks proof reading mechanism. The mutation rates in the order of 10-3has been reported for dengue (ElodieDes et al 2009) in different host settings. Such mutations often result in variants that become targets of selection; an outcome of underlying genotype and its environment. Despite these facts dengue virus do not evolve as fast as other RNA viruses. The only macro evolutionary divergence is perhaps the radiations in its four serotypes in its primate host (sylvatic strains) around one thousand years ago (ElodieDes et al 2009). There after genetic mutation in the envelope protein and receptor binding domains resulted in its emergence as infectious pathogen in human population. The divergent forms of these sylvatic strains are often found to be circulating in human habitat, suggesting that enzootic cycles with some spill over in the surrounding human population. This has been shown in Malaysian populations settled near forest and marshy habitats (Wang, E. et al., 2000). The phylogenetic studies conducted based on envelope gene sequences of basal portion of sylvatic linage, DENV 1,-2,-4 of Malaysian descent suggest that endemic / epidemic strains of these viruses diverged from sylvatic ancestors more than 1000 years ago (Wang, E. et al., 2000). Thereafter, only micro evolutionary change within dengue serotypes have taken place, these changes have nevertheless resulted in substantial genetic diversity with emergence of endemic and epidemic strains in different parts of the region.
Fig. 1. The effects of climatic and social change on vector evolution and disease severity
DEN-3 viruses have undergone independent evolution which has resulted in emergence of four genetic subtypes of which subtype I-III circulate in the South East Asian Region. Subtype I comprises of viruses from Indonesia, Malaysia and the Philippines; subtype II of viruses from Thailand and subtype III includes viruses from Sri Lanka India and Pakistan. The genetic evolution in these subtypes is primarily reported mutations in the prM/M and E structural protein genes. In spite of these mutations, the genomic region has retained greater than 95% amino acid sequence similarity (Lanciotti, R.S et al.,1994), suggesting that these are highly conserved regions responsible for protein architecture and / or biological function. Phylogenetic studies suggest that there are regional foci of virus extinction and selection, one such region is Thailand where the indigenous DEN-3 virus circulating up to 1992 disappeared and was replaced by two new lineages perhaps from a common ancestor (Wittke, V. et al. 2002). The sequence of all Thai DEN-3 isolates recovered after 1992 had T at position 2370 in contrast to the C at this site in the pre-1992 samples(Wittke, V. et al. 2002), and nucleotides difference was observed in at least 45 sites of total 96 sites studied. It appears that the post-1992 strains have replaced the pre-1992 strains). These studies point towards potential of regular extinctions of strains of DEN-3 virus and replacement by new variants in the region (Wittke, V. et al. 2002). Natural selection and / or genetic bottle neck could be the plausible causes for this variation. Since the extinction of pre 1992 strains and appearance of new epidemic strain in Thailand occurred during inter-epidemic period it is therefore hypothesized that the genetic bottleneck is perhaps the cause of regional replacement. This is further supported by studies from India reporting shift and dominance of the dengue virus serotype-3 (subtype III) replacing the earlier circulating serotype-2 (subtype IV) with emergence of increased incidence of DHF and DSS in subsequent outbreaks (Dash, P.K.et al. 2006). Strains from the 2005 outbreak in Karachi (Pakistan) were found to be similar to those from Indian strains of dengue serotype 3, and were responsible for deadly outbreak in 2005-06 (Jamil. B. et al. 2007). Thus over the period 1989 and 2000, a new clades of DENV-3 genotype III viruses have replaced older genotype and clades in this region and emergence of new clades coincided with severe epidemics. The epidemiologic data suggests that the DEN-3 virus responsible for recent epidemic outbreaks in Mozambiques, Gutamealla, Pakistan and SriLanka may have been introduced from India, and changing age structure of dengue patients from 1996-2005 may also be indicative of the selected virus moving into new areas(Kanakaratne, N. et al.2009 ).
Genetic evolution and disease severity in SEA
The micro evolutionary change within dengue serotypes has resulted in substantial genetic diversity with emergence of endemic and epidemic genotypes. With current advances in the field of genetic and molecular techniques scientists are now trying to decipher relation of changing clades with disease severity and epidemic potential. With the availability of complete genomic sequence of the Dengue virus different genetic loci have been investigated to find this relationship. Envelope -gene (E-gene) sequence is the most frequently investigated locus, (Wittke,V.et al., 2002;-Thu, H.M. et al.,2004;Islam, M.A.et al., 2006,27) followed by capsuler C-prM gene (Kukreti, H. 2008;,Dash, P.K. 2006;,Kanakaratne, N. 2009;Jamil B,2007). In addition non-structural (NS) viral proteins such as NS1 and untranslated genomic region 3′-UTR, 5′ UTR along with complete genomic sequences have been investigated to relate the genetic changes with the disease severity (Mangada, M.N. et al., 1997;Zhou,Y.et al.,2006;Islam, M.A.et al.,2006. Despite the wealth of genomic data available the exact cause and effect of viral virulence and clade changes is yet to be proven, however, viral linage is continually changing with local extinction and emergence of new clade. The introduction of new clade in the region translates in form of outbreaks of DHF and DSS. In order to analyze if there is a selection of specific clade in South East Asia that is circulating in the region and causing DHF outbreaks we conducted a meta-analysis. Studies conducted from 1950 to 2009 in South East Asian region that have investigated association of disease severity with specific sequence mutations in the dengue virus genome were retrieved. The objective was to analyze association of disease severity with the specific genomic mutation in the clade circulating and causing periodic epidemics in South East Asia. Since DENV-2 and DENV-3 are more common in this region our study was focused on these two genotypes only. Objectives of the metaanalysis were to identify association of specific genetic mutation in DENV-2 and DENV-3 with clinical severity seen during periodic epidemics in South East Asia. The specific review question was:Is clinical severity of dengue in the South East Asian region associated with emergence of specific mutations in genomes of DENV-2 and DENV-3 genotypes? We hypothesized that there is changing pattern of dengue virus genotypes in South East Asia and these mutations are associated with clinical severity of the disease.
Methods
Literature search
The literature search was performed from February 2010 to June 2010. Data sources include Medline via Pubmed (1950-February 2010), Cochrane data base of systematic reviews, Google scholar and experts in the field. Secondary references and review articles were scanned for thematic review. Hand search of the journal was also carried out. However, unpublished and ongoing studies could not be explored. Terminologies i.e. dengue type 1-4, dengue fever, dengue hemorrhagic fever, genetic variation, sequence analysis, south East Asia were used individually as well as in various combinations. Two independent reviewers; reviewed the titles, abstracts and full text articles and selected potentially relevant studies based on inclusion criteria established prior to the literature search. Discrepancy between the reviewers were sought to reach on consensus in consultation with third reviewer. Those potentially irrelevant studies that were ultimately excluded are listed together with the reason for exclusion in Table 1.
|
Author’s name |
Why excluded? |
|
Araujo J.M.G. et al, (2009) |
Conducted in Brazil, (also include dengue strains from regions other than South east Asia) but country of origin of these isolates was not clear, Sequences were selected from gene bank. |
|
Lanciotti RS.et al, (1994) |
Conducted in USA, origin of isolates not clear (also include dengue strains from other geographical regions other than South east Asia) The DEN-3 viruses used in this study were obtained from the collection at the Division of Vector-Borne Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins, Colo., U.S.A. |
|
Soundravally R, et al (2007) |
Focused on host factors as cause of disease severity rather than on virus gene mutations |
|
Soundravally R., et al (2008 a) |
Focused on host factors as cause of disease severity rather than on virus gene mutations |
|
Soundravally R., et al, (2008b) |
Focused on host factors as cause of disease severity rather than on virus gene mutations |
|
Gibbons RV. et al, (2007) |
Does not give sequence analysis in detail |
Table 1. Features of the excluded studies
Inclusion criteria
Studies which reported dengue virus genotype (mutation / sequencing of viral genetic material) and clinical features of dengue fever patients were included.
Design of the studies
All type of observational studies i.e. case report, case series, surveys and descriptive cross-sectional studies which were focusing on genotype and clinical presentation of dengue patients were included in the review. Population: Population includes patients of dengue fever of all age groups. No age and sex restriction were applied. Outcome of interest: Difference in nucleotide and protein sequences were analyzed and compared according to geographical origin, the sampling period and the clinical presentation. Clinical severity of the disease is defined as presence of DF, DHF or DSS. Language: Only articles in English language were included in the review.
Exclusion criteria
All those studies focusing on dengue vector control, clinical trials on vaccines, clinical trials on drugs, pure prevalence or incidence, unusual case report or case series without genotype and studies conducted in countries other than south East Asian region were excluded.
Data extraction
Data extraction of the included studies was done by using structured data extraction form specifically made for the review. Data was extracted for country of origin, year of publication, clarification of objectives, type of study, its duration and setting, results on both genotype and clinical severity etc.
Data synthesis
A narrative data synthesis was carried out to show result summary of all included studies which include description of clinical features and genotype of dengue virus. However, meta-analysis could not be performed due to non availability of required data i.e. measure of strength of association. Hence, pooled effect of genetic variation on clinical severities among dengue patients could not be provided.
Quality assessment
According to Cochrane Collaboration’s recommendation, the quality of included studies have been assessed by using criterion which asses the quality of studies by focusing on study type, sample size calculation, clarity of objective, selection of cases, and internal validity of selected studies.
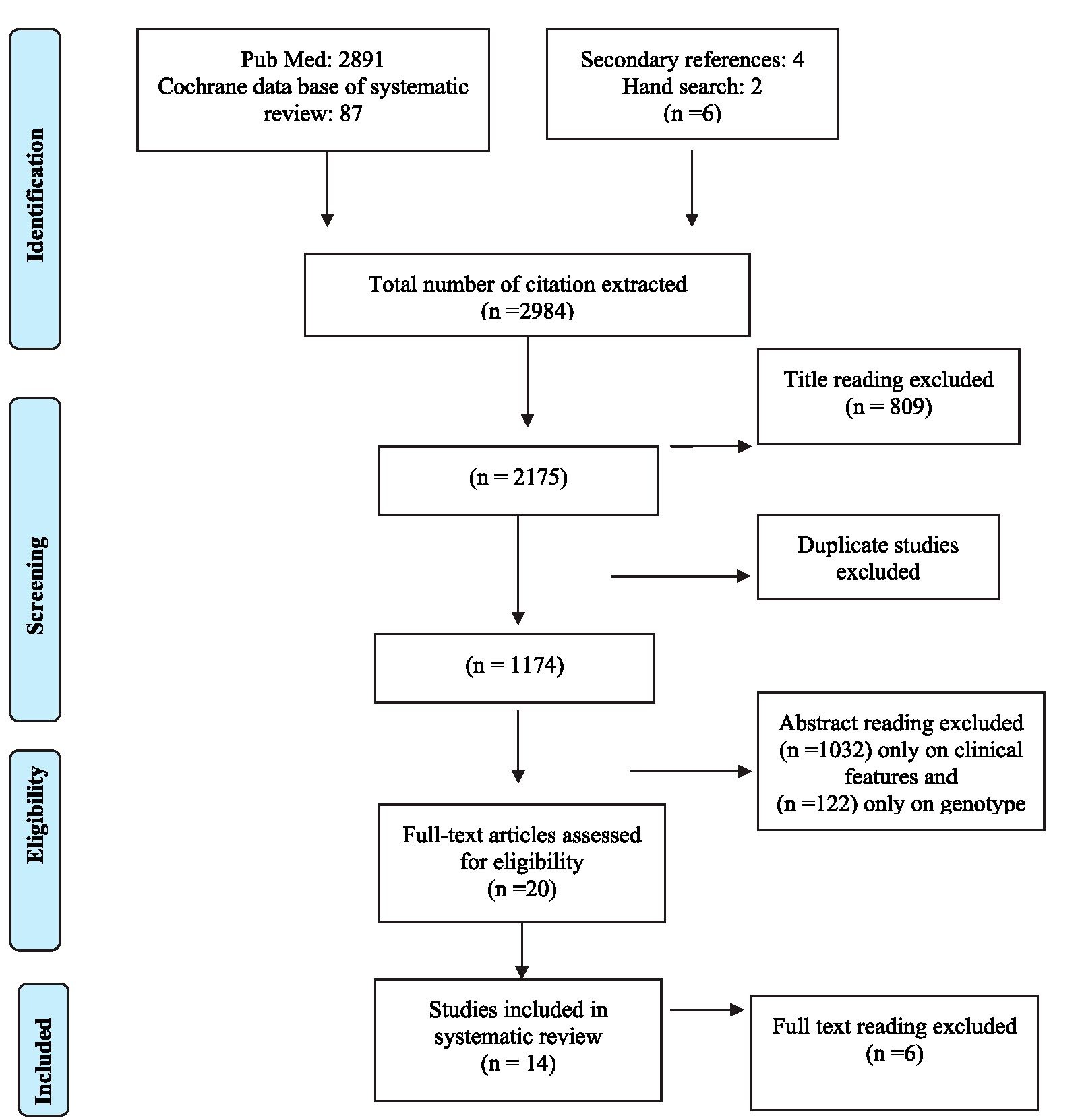
PRISMA Flow Diagram
Fourteen studies were finally selected based on inclusion criteria i.e., association of dengue genotype and clinical severity of the diseases in the patients and were conducted in different countries of South East Asia. Setting of these studies were; Thailand 6 (Zhang, C. et al 2006, Rico-Hesse R. et al. 1998, Wittke,V.et al.,2002), Myanmar (Thu H.M. et al.,2004), India (Kukreti, H. et al.,2008;Dash, P.K.et al.,2006), Bangladesh (Islam, M.A.et al.,2006) Sri Lanka (Kanakaratne, N. et al.,2009), Taiwan (King, C.C. et al.,2008) and Pakistan (Jamil B, et al 2007). These studies were published from 1997 to 2009. Since the focus of our study was on DEN-2 and DEN3 viruses 12 studies out of these 14 were finally included in this study
Study sample characteristics
Eight studies were conducted in the hospital settings (Zhang, C. et al 2006, Rico-Hesse R. et al. 1998, Wittke,V.et al.,2002, Kukreti, H. et a!.,2008;Dash, P.K.et al.,2006, Islam, M.A.et al.,2006, al,Kanakaratne, N. et al.,2009), three in the community (Zhou Y, et al 2006, Dash PK, et al 2006, Kanakaratne, N. et al.,2009), where as data was extracted from laboratory records in two studies (Jamil B, et al, 2007, King, C.C. et al.,2008) and dengue virus strain were taken from frozen stock of clinical serum samples in two (18, 21).
Age ranges for dengue patients in these studies varied from 1 year to 70 years. The total numbers of dengue patients were 7663 in these studies. Characteristics of the studies included in this review have been summarized in table 2. A total of 285 virus isolates were subjected to genotyping/ sequence analysis in these studies. All four genotypes were studied in three studies (Zhou Y, et al 2006,Jarman RG, et al 2008, Rico-Hesse R. et al. 1998); only DEN 3 in five studies (Wittke,V.et al.,2002, Kukreti, H. et al.,2008; Islam, M.A.et al.,2006), only DEN 2 in three studies (Zhou Y, et al 2006,Mangada MNM et al 1997, Zhang, C. et al 2006), only DEN 4 in one studies (KlungthongC, et al, 2004), whereas DEN 1 and DEN 3 in one study (Kukerti H, et al 2008) and DEN 1, DEN 2 and DEN 3 studied in one study (Jarman RG et al 2008).
Clinical definition
Dengue case was defined on the bases of presence of IgM, IgG, or fourfold or greater rise in hemagglutination inhibiting (HI) antibody titer against dengue virus, and presence of dengue virus specific nucleic acids in RT-PCR. Clinical severity was defined as presence of hemorrhagic manifestation and DHF related symptoms such as thrombocytopenia, skin rash, gum bleeding, gastrointestinal bleeding, hemorrhagic sclera, epistaxis, edema and ascitis. Where as other studies simply defined as presence of DF, DHF grade I, II and III and DSS as per WHO criteria.
Nucleotide sequencing and phylogenetic analysis
Envelope -gene (E-gene) sequence was most frequently investigated loci, nine studies were focused on this region followed by C-prM gene, in three studies both genetic loci studied in one study (9) and one study included NS1 along with PrM and E loci. The 3′-UTR, 5′ and 3′ UTR and complete genomic sequences were studied in one each
Homology search and comparisons of most obtained sequences were performed using commercially available software systems such as DNASIS, DNAStar, 3′ -UTR secondary structures were estimated using MFOLD package, while nucleotide sequence alignments (Phylogenetic analysis) were performed using CLUSTAL X, MEGA version, and maximum likely hood methods available e.g. PAUP PROGRM
The quality of included studies was assessed by using criterion which asses the quality of studies by focusing on study type, sample size calculation, clarity of objective, selection of cases, and internal validity of selected studies. From total of 16 points scale, individual score on quality assessment criteria was as follows 8.5 (Rico-Hesse R. et al. 1998), 7.0 (Zhou Y, et al 2006,Jarman RG, et al 2008, Jamil B, et al, 2007), 10.5 (Dash PK, et al 2006, Kukreti, H. et al.,2008), 5.0 (Wittke,V.et al.,2002), 6.5 (Zhang, C. et al 2006,), 10 (Zhou Y, et al 2006, Mangada MNM et al 1997), 7.5 (Wittke,V.et al.,2002, Jarman RG, et al 2008, King, C.C. et al.,2008), 12(KlungthongC, et al, 2004). Since most of the severe DHF outbreaks in SEA have been associated with DEN-2 and DEN-3, mutational changes and its relation to disease severity of these two serotypes will be discussed here in detail.