1. Introduction
The recent advances in analyses of completely sequenced genomes of numerous model organisms, including the human genome, have revealed that approximately one-third of all predicted gene products of a given organism are likely to be associated with membranes (Auerbach et al., 2002). These integral and membrane-associated proteins execute a variety of essential cellular tasks. For example, transmembrane receptors bind specific signaling molecules such as hormones, growth factors, and neurotransmitters, and initiate specific cellular responses. Ion channels, transporters, and pumps mediate the exchange of membrane-impermeable molecules between cellular compartments, and between a cell and its extracellular environment. Cell-adhesion molecules enable many animal cells to adhere tightly and specifically with cells of the same or similar type ensuring their segregation into distinct tissues (Fetchko et al., 2003). Because of their exposure to the extracellular space, membrane proteins are also of considerable pharmacological importance. Namely, 50% of currently known drug targets (~500) are either membrane receptors or ion channels. Altogether, the central role of membrane proteins in many cellular processes, their direct link to human disease, and their accessibility toward drugs makes an understanding of membrane protein function desirable.
However, the study of membrane proteins has historically been problematic. The hydrophobic nature of membrane proteins often results in insoluble proteins, which makes protein isolation difficult and therefore hinders the determination of protein complex composition and protein function. Recently, several yeast genetic techniques have made the characterization of interactions among membrane proteins more feasible (Reviewed in (Auerbach et al., 2002)). By characterizing protein interactions, researchers can link unknown proteins to proteins of known function, thus providing insight into the unknown protein function. Also, since protein interactions are central in regulating protein activity, knowledge of protein interactions provides understanding of both normal and mutant protein activity.
This review provides a brief overview of the available genetic methods for detecting protein interactions among membrane proteins.
2. Technologies for detecting membrane protein interactions
Generally speaking, the approaches for dissecting protein-protein interactions between membrane proteins can be divided into biochemical and genetic methods (Stagljar, 2003). Biochemical methods establish the state of interacting proteins by directly working with proteins in order to determine the composition of protein complexes, whereas genetic methods indirectly determine protein interactions on the basis of outputs produced through the manipulation of endogenous or exogenous gene networks. Traditionally, biochemical methods such as copurification or coimmunoprecipitation have been used to investigate the composition of protein complexes (Adams et al., 2002; Einarson and Orlinich, 2002; Pedrazzi and Stagljar, 2004). However, these methods are tedious and time consuming, requiring extensive optimization for each given protein pair and are therefore unsuitable for the simultaneous application to the tens of thousands of uncharacterized proteins predicted from genome sequences.
Aside from biochemical approaches, expression systems that are based on the detection of protein-protein interactions in vivo have become a popular tool because they require little individual optimization and are well suited for screenings in a high-throughput format. Among genetic assays, the YTH system takes a special position (Fields and Song, 1989). The organism used for the YTH system, the unicellular eukaryote Saccharomyces cerevisiae, is well suited to high-throughput screenings, is thoroughly characterized (completely sequenced genome), easy to handle, grows quickly, is well suited for the expression of heterologous proteins, and contains a multitude of selective markers available for nutritional selection (e.g., HIS3, URA3, TRP1, LEU2), drug susceptibility (e.g., URA3, cyhR), drug resistance (e.g., kanMX, natMX), and colors (e.g., LacZ, GFP, GusA) (Melese and Hieter, 2002).
Despite the fact that YTH is a powerful and versatile technique to identify new partners, this method is limited because it cannot be applied to all proteins. For example, since in the YTH system one reconstitutes and monitors TF activity, all assays must be performed within the nucleus. This situation presents a problem when studying membrane proteins, since, in order to evaluate membrane protein interactions by the YTH method, the membrane protein must be portioned into fragments and then relocalized to the nucleus. This limitation not only affects the presentation of the protein of interest, but also limits the protein interacting partners, which can be detected by the YTH system as some membrane proteins cannot be effectively expressed in the nuclear milieu. In addition, membrane proteins are often co- and posttranslationaly modified (they are glycosylated and/or acquire disulfide bridges), and these modifications are not expected to occur in the nucleus (Thaminy and Stagljar, 2002). To overcome these problems, several yeast-based strategies have been designed to study protein interactions in their natural environment -membrane (Stagljar and Fields, 2002).
2.1. G-protein fusion assay
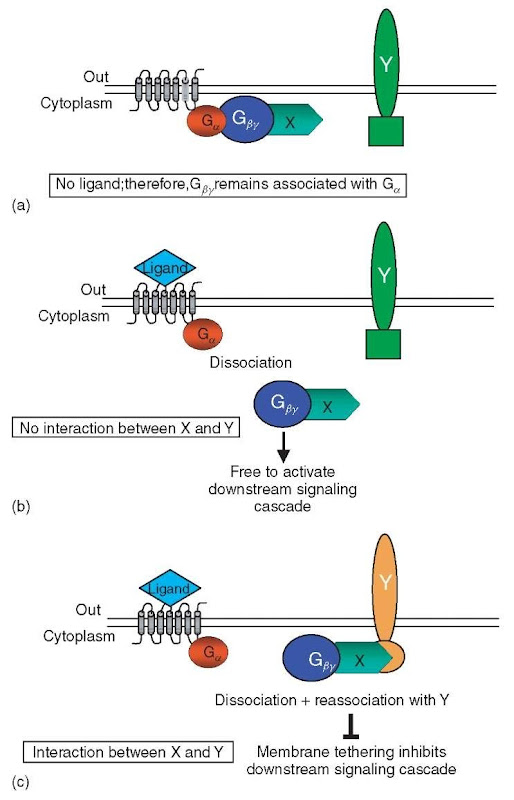
The G-protein fusion assay utilizes the G-protein coupled yeast mating pathway receptor to study protein interactions among membrane proteins (Ehrhard et al., 2000). G-protein coupled receptors are composed of three subunits, Ga, G^, and GY (Figure 1). When the Ga subunit of the G-protein coupled receptor binds a ligand (either mating factor a or a), GTP is exchanged for GDP on Ga, which leads to the dissociation of the trimeric receptor complex into Ga and GpY. Once the G^y subunits are released from Ga, GpY is no longer tethered to the membrane, which enables it to act on various downstream effectors thus activating the mating cascade.
In order to detect membrane protein interactions, the yeast-mating pathway G-protein signaling process was modified in the following manner. An unmodified integral membrane protein-Y serves as a “bait” and a “prey” X is fused to the G^y subunit of the G-protein coupled receptor (GpY -X) (Figure 1). In the presence of ligand, if no interaction between GpY -X and the integral membrane protein Y occurs, the activation of the mating cascade is stimulated and is either detected by an indirect transcriptional readout or by a characteristic cell morphology change, that is, Schmoo formation, induced in the presence of the mating factor ligand (Figure 1b). However, if protein GpY -X interacts with Y, the Y integral membrane protein continues to sequester GpY -X at the membrane, which blocks the downstream mating cascade (Figure 1c).
The G-protein fusion assay has been shown to detect known interactions between the binding partners syntaxin 1a and neuronal Secl (nSecl), and between the fibroblast-derived growth factor receptor 3 (FGFR3) and SNT-1 (Ehrhard et al., 2000). In addition, mutations of nSec1 that fail to bind syntaxin 1a were isolated using the G-protein fusion assay (Ehrhard et al., 2000).
2.2. Reverse Ras recruitment system (reverse RRS)
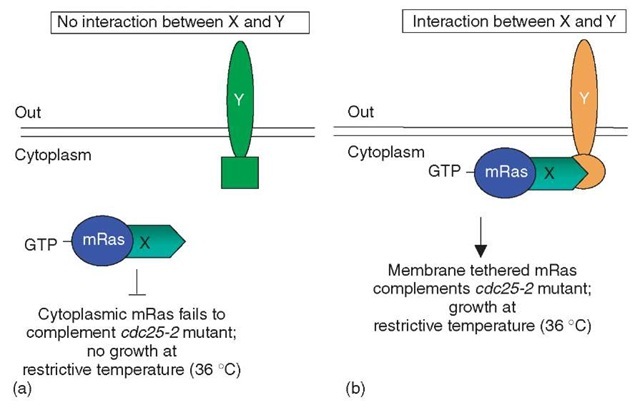
In yeast, activation of Ras via the exchange of bound GDP for GTP is mediated by the Cdc25 guanyl exchange factor and results in cell growth. Temperature-sensitive mutations of cdc25 (allele- cdc25-2) grow normally at the permissive temperature of 23°C but fail to grow at the restrictive temperature of 36°C. A constitutively active cytoplasmic version of a mammalian Ras (mRas), which lacks the carboxy-terminal domain “CAAX box” required for normal plasma membrane association, is unable to complement the growth phenotype of the cdc25-2 allele at 36°C since mRas is not at the plasma membrane (PM). However, if the constitutively active cytoplasmic mRas is associated with the PM via protein-protein interactions, the growth phenotype of the cdc25-2 mutant yeast at 36°C is complemented. In the reverse RRS, an unmodified integral membrane protein Y “bait” is expressed together with a “prey” protein X fused to mRas (X-mRas) within a cdc25-2 mutant yeast strain (Hubsman et al., 2001) (Figure 2). If Y and X-mRas fail to interact, no growth of cdc25-2 mutants will be observed at 36° C since X-mRas is not targeted to the PM (Figure 2a). However, if Y and X-mRas interact, X-mRas is recruited to the PM, thus allowing the growth of cdc25-2 mutant cells at the restrictive temperature of 36°C (Figure 2b).
The reverse RRS has been used to isolate two novel interacting partners of the small GTPase Chp (Hubsman et al., 2001). Also, through the use of promoters expressing different levels of the X-mRas prey, Kohler and Muller were able to detect interactions between two membrane-associated proteins (Kohler and Muller, 2003). In this case, high to moderate expression levels of the membrane-associated prey, the adaptor protein growth factor receptor binding protein 2 (Grb2) fused to a myristoylation signal and mRas, supported only slight growth of the cdc25-2 mutant yeast. Upon addition of an interacting bait, substantial increases in cell growth were observed presumably from the enhancement or stabilization of the X-mRas membrane localization, or perhaps clustering of the Ras facilitated further signaling (Kohler and Muller, 2003). Either way, for certain cases the limitation of using membrane associated proteins as preys, which normally would result in cell growth in the absence of protein interactions, has been eliminated. However, it is still possible that the Grb2 bait used by Kohler and Muller is a special case since Grb2 is a member of the Ras signaling pathway. Furthermore, the question still remains whether the RRS can be used to examine integral membrane proteins as baits.
Figure 1 G-protein fusion assay: The G-protein fusion assay can test for interactions between a native integral membrane protein Y and a protein X, which is fused to the Gpy subunit of the G-protein coupled receptor (G^-X). (a) When no ligand is present, the Gpy subunit of the G-protein coupled receptor (Gpy-X) remains associated with the Ga subunit of the G-protein coupled receptor and no mating cascade events are activated. (b) If no interaction occurs between Y and Gpy-X, upon ligand binding of the receptor, the Gpy -X subunit dissociates from Ga and therefore is free to activate downstream signaling events. (c) If an interaction occurs between Y and GpY -X, the integral membrane protein Y sequesters Gpy-X at the membrane, which in effect inhibits downstream signaling events although in the presence of ligand
Figure 2 Reverse Ras recruitment system: The Reverse Ras recruitment system tests for interactions between a cytoplasmic constitutively active mRas, which is fused to protein X (X-mRas), and a membrane associated protein Y. (a) If no interaction occurs between proteins X-mRas and Y, protein X-mRas remains cytoplasmic and is unable to complement the growth phenotype of the mutant yeast strain cdc25-2. (b) However, if X-mRas and Y interact, X-mRas is then tethered to the membrane, which enables X-mRas to complement the growth phenotype of the mutant yeast strain cdc25-2
2.3. Split-ubiquitin assay systems
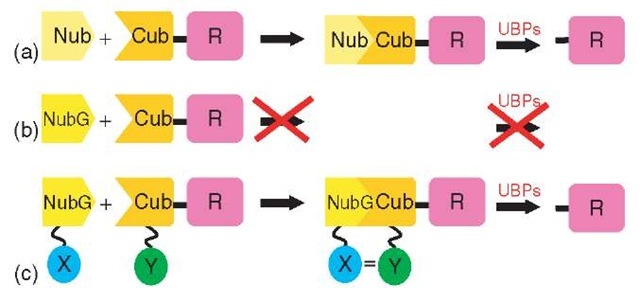
The split-ubiquitin system provides another approach to study membrane protein interactions (Stagljar et al., 1998; Wittke et al., 1999). Ubiquitin (Ub) is a small, highly conserved protein that is attached to lysine residues of proteins in order to tag the proteins for proteasomal degradation (Hershko and Ciechanover, 1992). Ubiquitin-tagged proteins are recognized by ubiquitin-specific proteases (UBPs) that cleave after the C-terminal (Gly 76) residue of Ub and the first residue of the target protein, allowing release of the protein for degradation by the 26 S proteasome. Johnsson and Varshavsky found that native ubiquitin can be split into an N-terminal (Nub) and a C-terminal (Cub) half (Johnsson and Varshavsky, 1994). The two halves retain a basic affinity for each other and spontaneously reassemble to form quasi-native ubiquitin. If a reporter protein is fused to the C-terminus of Cub, it will be cleaved off by UBPs upon assembly of the Nub and Cub moieties (Figure 3a). A point mutation in the N-terminal domain of ubiquitin (NubG) abolishes the affinity of the two halves for each other, such that NubG and Cub fail to refold into split-ubiquitin when coexpressed in yeast (Figure 3b). However, if the two ubiquitin halves are fused to the interacting proteins X and Y, this interaction brings the NubG and Cub moieties close enough together to reconstitute quasi-native Ub, resulting in the release of the reporter protein by the UBPs (Figure 3c).
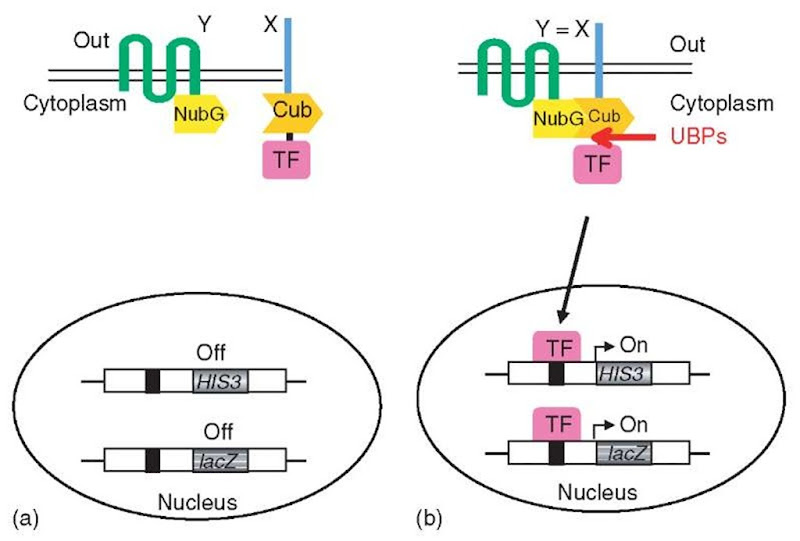
The versatility of transcriptional activation of reporter genes in yeast was used to convert the split-ubiquitin system into a genetic assay for the in vivo detection of membrane protein interactions (Stagljar et al., 1998). In this membrane-based YTH assay (Figure 4), an artificial transcription factor (TF) consisting of the bacterial LexA protein and the Herpes simplex VP16 transactivator protein is fused to the Cub moiety. An integral membrane protein (X) is expressed as a fusion to the Cub-LexA-VP16 reporter cassette, with this cassette attached either to the Nor C-terminus of the transmembrane protein, depending on the orientation in the membrane of this protein. The second protein under investigation (Y), either another transmembrane protein or a cytoplasmic protein, is expressed as a fusion to NubG. If interaction between the X and Y proteins occurs, a split-ubiquitin molecule can be reconstituted, leading to the proteolytic release of the transcription factor to activate a reporter gene (Stagljar et al., 1998). Thus, the reassociation event initiated by the protein interaction is converted into a transcriptional output that can be easily detected. This assay has been used to investigate the influence of mutations on the assembly of fragments of presenilin (a protein implicated in the Alzheimer’s disease) (Cervantes et al., 2001), to characterize the interaction between the yeast a1,2-mannosidase Mns1p and Rer1p (Massaad and Herscovics, 2001) and Wbp1p and Sss1p in the endoplasmic reticulum (Scheper et al., 2003), and to study intra-and intermolecular interactions between plant sucrose transporters (Reinders et al., 2002). This approach has been successfully adapted for prey library screening and has identified three novel interacting partners of mammalian ErbB3, a receptor tyrosine kinase involved in regulation of proliferation and differentiation of many tissue types (Thaminy et al., 2003), and BAP31, a human polytopic integral membrane protein of the endoplasmic reticulum involved in apoptosis (Wang et al., 2003). Thus, MbYTH allows rapid and sensitive characterization of proteins associated with a particular full-length transmembrane protein of interest and is generally applicable to most transmembrane signaling proteins.
Figure 3 Split-ubiquitin: (a) Ubiquitin can be expressed as an N-terminal (Nub) half as well as a C-terminal (Cub) half that is fused to a reporter protein. The two halves retain affinity for each other and spontaneously reassemble to form the so-called split-ubiquitin. (b) A point mutation in the N-terminal half of ubiquitin (NubG) completely abolishes the affinity of the two halves for each other, and as the separate NubG and Cub parts are not recognized by ubiquitin-specific proteases (UBPs), no detectable cleavage of the attached reporter takes place. (c) NubG and Cub are fused to the interacting proteins X and Y. The X-Y interaction brings the NubG and Cub domains close enough together to reconstitute ubiquitin, resulting in the release of the reporter protein by the action of the UBPs
Figure 4 The split-ubiquitin membrane YTH system (MbYTH): (a) A membrane protein of interest X is fused to Cub followed by an artificial transcription factor (TF), while another membrane (or cytoplasmic) protein Y is fused to the NubG domain (Y-NubG). Co-expression of X-Cub-TF with a noninteracting Y-NubG does not lead to the formation of split-ubiquitin nor cleavage by UBPs. (b) On interaction of the X and Y proteins, ubiquitin reconstitution occurs, leading to proteolytic cleavage and the subsequent release of the transcription factor. This factor activates reporter genes to result in cells that are histidine prototrophs and that turn blue in a f-galactosidase assay
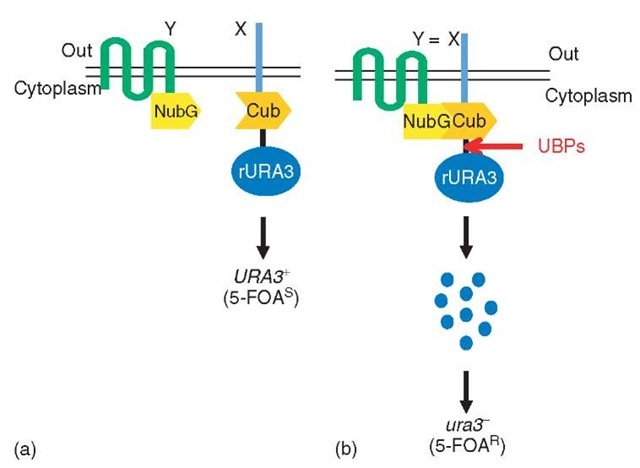
In another split-ubiquitin-based approach, Johnsson, and colleagues (Wittke et al., 1999) have fused a destabilized version of the yeast Ura3 protein, termed rUra3, to the Cub moiety. An integral membrane protein X is expressed as a fusion to the Cub-rUra3 cassette. If cells expressing the fusion protein are grown on a medium containing the compound 5-fluoro-orotic acid (5-FOA), they die because the rUra3 protein converts 5-FOA into a toxic product. However, if the cells coexpress an interacting protein Y fused to NubG, the Cub, and NubG moieties can be forced into close proximity by the X-Y interaction and associate to form split-ubiquitin. This association, in turn, leads to UBP-mediated cleavage at the C-terminus of Cub and the release of the rUra3 fusion protein into the cytosol. Since the newly created N-terminal residue of the rUra3 protein is destabilizing in the N-end rule pathway of protein degradation (Varshavsky, 1997), the entire fusion protein is degraded by the 26 S proteasome, leading to cells that can grow on medium containing 5-FOA (Wittke et al., 1999). In this way, cells expressing two interacting proteins can be identified by their ability to survive selection on 5-FOA plates (Figure 5). The rUra3-based split-ubiquitin method was used to map the interactions between several S. cerevisiae integral membrane proteins (Wittke et al., 1999) and to analyze changes in protein conformation and stability of the S. cerevisiae protein Sec62, a component of the translocation machinery in the membrane of the endoplasmic reticulum (Raquet et al., 2001). In addition, the system has recently been used to systematically test the pairwise interactions between peroxins, proteins that play a role in protein import in the peroxisomes (Eckert and Johnsson, 2003).
Figure 5 The rUra3-based membrane YTH system: (a) A membrane protein (X) under investigation is expressed as a fusion to the Cub domain, which is fused to a destabilized version of the Ura3 protein (rUra3). The NubG domain is linked to the membrane protein Y. If X and Y do not interact, there is no ubiquitin reconstitution and thus no UBP-mediated cleavage, resulting in yeast cells that contain Ura3 activity and thus die (5-FOAS) on medium containing 5-fluoro-orotic acid (5-FOA), a toxic metabolite of the Ura3 enzyme. (b) If the X and Y proteins interact, the Cub and NubG domains are brought into close proximity, where they reconstitute an active ubiquitin. Cleavage by UBPs then releases rUra3 from the Cub fusion. The cleaved rUra3 is targeted for rapid destruction by the enzymes of the N-end rule to yield cells that are uracil auxotrophs (ura3) and 5-FOA resistant (5-FOAR)
3. Conclusions
Membrane-associated proteins perform a critical role in many essential cellular processes and thus represent one of the most important class of drug targets; currently, they account for ~50% of all known pharmaceutical drug targets. However, analysis of membrane protein interactions is notoriously difficult because of the hydrophobic nature of such proteins, which makes them unsuitable bait candidates to use in most of the interactive proteomic technologies. The development of TAP technology linked to mass spectrometry analyses as well as the YTH system has provided a valuable means to identify proteins that physically interact in vivo (Fields and Sternglanz, 1994; Rigaut et al., 1999). Furthermore, in the past four years, the scientific community has witnessed the development of several genetic assays in yeast that are capable of detecting membrane protein interactions (Ehrhard et al., 2000; Hubsman et al., 2001; Stagljar et al., 1998; Urech et al., 2003; Wittke et al., 1999). In addition, genetic assays based on complementation of proteins or protein fragments have also been developed in organisms other than yeast that allow the monitoring of membrane protein interactions in real time, including ones amenable to use in mammalian cells (Blakely et al., 2000; Remy and Michnick, 1999; Rossi et al., 1997). Altogether, these assays represent an important step forward toward the elucidation of physical protein-protein interactions, since prior to their development the interactions of membrane proteins were difficult to study using the then existing biochemical and genetic methods. In the near future, a logical extension of the use of various such interactive proteomic technologies for membrane proteins will be their adaptation in a high-throughput format. This approach, in combination with automated screens, should help in elucidating membrane protein interactions on a genome-wide scale. Clearly, such approach will be highly instructive in understanding the pathways underlying many human diseases and will most likely reveal new targets for their therapy. Lastly, such knowledge can be used to provide predictors of certain diseases and may thus be used to develop biomarkers as fingerprints for particular disorders or cellular responses.