1. Introduction
Two-dimensional polyacrylamide gel electrophoresis (2D PAGE) has been widely used over the past four decades to resolve several thousand proteins in a single sample. This has enabled the identification of the major proteins in a tissue or subcellular fraction by mass spectrometric methods. In addition, 2D PAGE has been used to compare relative abundances of proteins in related samples, such as between mutant and wild type organisms or control and diseased tissues, allowing the response of classes of proteins to be determined. To date, the majority of comparative protein-profiling studies have produced qualitative data, which have enabled the investigator to determine whether or not a particular protein shows an increase or decrease in expression. This provides no measure of the extent of this change in expression, therefore, it is unsuitable for the clustered data analysis needed for an insight into functionality. Quantitative proteomics allows coexpres-sion patterns to be studied, and proteins showing similar expression trends may be assigned membership of the same functional groups; however, quantitation has been hampered by several factors. Firstly, silver staining, being more sensitive than Coomassie staining methods, has been widely used for high-sensitivity protein visualization on 2D PAGE, but it is unsuitable for quantitative analysis as it has a limited dynamic range. The most sensitive silver staining methods are also incompatible with protein-identification methods based on mass spectrometry. More recently, the Sypro family of postelectrophoretic fluorescent stains (Molecular Probes, Eugene, Oregon, USA) have emerged as alternatives, offering a better dynamic range than silver staining and ease of use (Malone et al., 2001). Another problem is the irreproducibility of 2D gels; no two gels run identically meaning that corresponding spots between two gels have to be matched prior to quantification. Finally, normalization has proved challenging, especially in the case of silver staining where staining is protein dependent. These aforementioned factors all add variability to the system that make it unsuitable for the accurate quantitation.
Difference gel electrophoresis (DIGE) circumvents many of the issues associated with traditional 2D PAGE, such as gel-to-gel variation and limited dynamic range, and allows more accurate and sensitive quantitative proteomics studies. This minireview is an overview of this technique, describing its strengths and limitations.
The DIGE technique was first described some time ago by Jon Minden’s laboratory (Unlu et al., 1997) and is now available as a technique from Amersham Biosciences. This technique relies on preelectrophoretic labeling of samples with one of three spectrally resolvable fluorescent CyDyes (Cy2, Cy3, and Cy5), allowing multiplexing of samples into the same gel. There are currently two types of CyDye labeling chemistries available from Amersham Biosciences.
2. Minimal labeling
The most established chemistry is employed in the “minimal labeling” method, which has been available from this supplier since July 2002. Here, the CyDyes are supplied with an N -hydroxy succinimidyl ester group that reacts with the epsilon amino group of lysine side chains. Labeling reactions are engineered such that the stoichiometry of protein to fluor results in only 2-5% of the total number of lysine residues being labeled. It is imperative to keep a low dye:protein ratio to avoid multiple dye additions, as these would result in multiple spots being resolved in the second dimension of the DIGE gel. The typically high lysine content of most proteins makes it challenging to force the labeling reaction to saturation without using excessive amounts of reagent. The fluors carry an intrinsic charge of +1, such that the pI of the protein is preserved upon labeling. The three fluors are also mass matched, each labeling event adding approximately 500 Da to the mass of the protein. Labeling with CyDye DIGE Fluors is very sensitive with a detection limit of around 500 pg of a single protein and a linear response in protein concentration over at least five orders of magnitude. In comparison, the limit of detection with silver stain is in the region of 1ng of protein with a dynamic range of no more than two orders of magnitude (Lilley et al., 2002; Tonge et al., 2001). The labeling system is compatible with the downstream processing commonly used to identify proteins, which involves the generation of tryptic peptides. Trypsin cleaves the peptide bonds on the C-terminal side of lysine and arginine residues, but, as so few lysine residues are modified by dye labeling, peptide generation is largely unhindered. It is also unlikely that a peptide modified with a CyDye DIGE Fluor would be extracted from a gel piece, hence, interference by the fluors in peptide mass fingerprinting and de novo sequencing mass spectrometric techniques is minimal. A drawback of this minimal labeling system is the fact that the majority of the protein within a sample remains unlabeled and, in the case of smaller molecular weight species, the labeled portion of the protein may migrate to a slightly different position on a 2D gel. To ensure that the maximum amount of protein is excised for downstream processing, minimally labeled DIGE gels are often poststained with a total protein stain such as SyproRuby.
3. Saturation labeling
The second more recent chemistry was released by Amersham Biosciences in July 2003 and is designed for use in situations where sample abundance is limited. This differs from the original N -hydroxy succinimidyl chemistry in that CyDyes with no intrinsic charge are supplied with a thiol-reactive maleimide group. These “saturation” dyes are utilized in such a way to bring about labeling of every cysteine residue within a protein. The saturation labeling is much more sensitive as more fluorophor is introduced into each protein species, Shaw et al. reporting an order of magnitude increase in sensitivity over the original minimal dyes (Shaw etal., 2003). While the added sensitivity that these dyes provide is desirable, their use is technically more challenging. The reaction conditions have to be carefully optimized for each type of sample to ensure complete reduction of cysteine residues and a protein:dye ratio sufficient for stoichiometric labeling. Substoichiometric labeling will lead to multiple spots in the second dimension, whereas the use of too much dye may lead to unwanted addition reactions with lysine residues, resulting in the formation of charge trains in the first dimension. It is also impossible to compare the 2D spot maps between samples labeled with the two chemistries. Proteins containing multiple cysteine residues may appear as larger molecular weight species when labeled with the saturation dyes. For studies where identification by mass spectrometric techniques is required, a preparative gel with increased protein loading will be required to produce a 2D protein map. In situations where the amount of sample is very small, this technique at best can be considered of diagnostic value. The use of these saturation dyes is not well established, and the maleimide dyes are currently only available for Cy3 and Cy5.
4. Experimental design
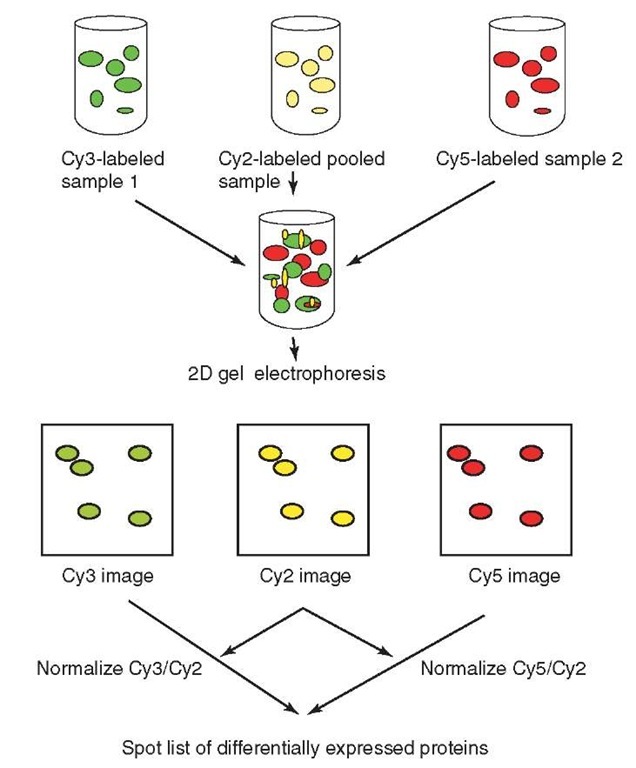
DIGE is a particularly powerful approach to study the changes in protein abundance in experimental studies involving comparison of multiple samples. In such studies involving the use of minimal dyes, it is desirable to label samples with either Cy3 or Cy5 minimal dyes, whereas Cy2 is used to label a pooled sample comprising equal amounts of each of the samples within the study, and acts as an internal standard (see Figure 1). This ensures that all proteins present in the samples are represented, allowing both inter- and intra-gel matching. Variation in spot volumes due to gel-specific experimental factors, for example protein loss during sample entry into the immobilized pH gradient strip, will be the same for each sample within a single gel. Consequently, the relative amount of a protein in a gel in one sample compared with another will be unaffected. The spot volumes are normalized for dye discrepancy, arising from differences in laser intensities, fluorescence, and filter transmittance, using a method based on the assumption that the majority of protein spots have not changed in expression level (Alban et al., 2003). The spot volumes from the labeled samples are compared to the internal standard giving standardized abundances. This allows the variation in spot running success to be taken into consideration. For the analysis, software developed for the DIGE system (DeCyder™ by Amersham Bioscience, Sweden) is typically used. This software has a codetection algorithm that simultaneously detects labeled protein spots from images that arise from the same gel and increases accuracy in the quantification of standardized abundance (Alban et al., 2003). The standardized abundances can then be compared across groups to detect changes in protein expression. In the case of the saturation dyes, where a Cy2 label is not available, the internal standard is labeled with one of the dyes and individual samples appear on separate gels labeled with the other dyes. This approach reduces the extent of multiplexing and increases the number of gels within a comparative experiment (Shaw et al., 2003).
Figure 1 Schematic outline of a 2D DIGE study using an internal pooled standard constructed from equal amounts of all the samples in the study, labeled with Cy2. Samples 1 and 2 are labeled with either Cy3 or Cy5. Each 2D gel performed within the study will have the sample Cy2 standard, combined with the Cy3 and Cy5 labeled samples prior to electrophoresis. The spot intensities from samples 1 and 2 can be normalized using the corresponding Cy2 spot intensities. This approach allows the measurement of more subtle protein expressional differences with increased statistical confidence
The design of a protein-profiling analysis experiment using DIGE is crucial to the amount of statistical significance that can be placed on the data. Consideration must be given to methods employed to assess both biological and experimental noise within the system being studied and ample biological and technical replicates must be processed. It is essential to measure the experimental variation in the DIGE process for any new set of samples. This can be achieved by running sets of gels where the same sample is labeled with all dyes to be used, loaded onto one gel, and fully analyzed. This will also give an indication of the inherent error in the system and suggest the threshold of significance or a fold change above which true changes in expression can be measured. In the case of the minimal dyes, a system bias at low spot volumes is observed owing to the different fluorescence characteristics of acrylamide at the different wavelengths of excitation for each of Cy2, Cy3, and Cy5. This system bias can be greatly reduced by employing a dye swap approach; several replicate gels are run where each sample appears with the opposite labeling, that is, gel 1 would contain pooled sample in Cy2 channel, wild type in Cy3 channel, and mutant in Cy5; gel 2 would contain pooled sample in Cy2 channel, mutant in the Cy3 channel, and wild type in Cy5 channel (Karp et al., 2004). The multigel approach allows many data points to be collected for each group to be compared. Spots of interest can be selected by looking for significant change across the groups, for example, with a univariate statistical test, for example, Student’s t-test or analysis of variance (ANOVA). These give a probability score (p) for each spot. This score indicates the probability that the groups are the same, consequently a spot with a low score, for example, p < 0.05, represents significant difference in relative abundance. The number of replicates required in a study depends on the amount of variation in the system being investigated and on how small the changes in expression are that you wish to measure at a given confidence level. Increasing the number of replicates will increase confidence in smaller changes in expression.
5. Drawbacks of the system
Regardless of the benefits of DIGE, the 2D PAGE process itself has some limitations. For global expression analysis, every protein should be resolved as a discrete detectable spot, The following groups of proteins, however, are often poorly represented: those with extreme isoelectric points (pI) or molecular weight; hydrophobic proteins; lower abundance proteins. It has been calculated that somewhere in the region of 90% of the total protein of a typical cell is made up of only 10% of the 10 000-20 000 possible different species, and hence many low abundance proteins may not be detectable (Zuo etal., 2001). Co-migration is also an issue, with proteins of similar pI and denatured molecular weight becoming focused at the same position of the gel. This makes it impossible to accurately determine the relative abundance of an individual protein within a mixed spot. There continues to be improvements to the 2D PAGE technique, however. Enhanced resolution of protein species can be achieved by the use of narrow range immobilized pH gradient (IPG) strips and/or prefractionation of sample; these greatly improve the chance of identification and assignment of function to scarce species (Tonella etal., 2001). Membrane proteins remain a problem. The use of more rigorous detergents such as amidosulphobetaine 14 (ASB14) has increased the number of membrane-associated proteins, which can be resolved by 2D PAGE (Santoni et al., 2000). In the case of studies involving integral membrane proteins, a 2D PAGE approach should be avoided and differential isotopic labeling strategies involving protein digestion in solution digestion to peptides employed (Li etal., 2003).
One of the main criticisms of DIGE is the financial outlay necessary to install the system in a laboratory. An appropriate scanning system and dedicated software are required and the cost of CyDyes necessary for large scale studies is not trivial.
6. Applications of DIGE
To date, the DIGE technology has been used with great success to study a variety of systems, allowing the detection of more subtle changes in protein expression than conventional methods where separate samples are loaded onto each gel (Gade et al., 2003); these include breast cancer cells (Gharbi et al., 2002), cat brain (Bergh et al., 2003), esophageal cancer cells (Zhou et al., 2002), yeast (Hu et al., 2003), chloro-plast (Kubis et al., 2003), GPI-anchored proteins (Borner et al., 2003), murine mitochondria (Kernec et al., 2001), mouse brain (Skynner et al., 2002), and rat heart (Sakai et al., 2003), although as yet there is a dearth of publications where the internal standard system is used. Knowles et al. (2003) have described a significant increase in the accuracy of determination of differential protein expression using the Cy2-labeled internal standard approach. They compared the relative abundances of proteins in cerebral cortex from wild-type mice and neurokinin 1 receptor knockout mice to elucidate molecular pathways involving this protein. They also compared relative abundances and significance values for differentially expressed spots derived from gels incorporating the pooled Cy2-labeled standard with values derived from the same gels, but without normalizing spot volumes to the corresponding pooled standard. They demonstrated that virtually all differentially expressed spots gave lower significance levels and a higher incidence of false-positives when derived without using the pooled standard for normalization. The authors reported being able to measure as little as 10% change in abundance with 95% confidence (p < 0.05).
In conclusion, 2D DIGE is the most powerful 2D PAGE-based approach for widespread protein-profiling studies by virtue of the ability to multiplex and link samples across numerous different gels in a study using an internal standard. This approach also gives information about more subtle changes in protein expression than conventional 2D PAGE. The advent of the saturation dyes has increased the sensitivity of this system. The use of 2D DIGE, however, will not result in a global analysis of a proteome as membrane proteins and proteins with extremes of pI and molecular weight will be poorly represented. In this respect, other quantitative techniques such as differential labeling with stable isotope can be considered to be complementary.