abstract
In this article we report upon our experiences of developing Web-services based infrastructures within two e-health projects. The first—a small demonstrator project funded by the UK’s National Cancer Research Institute (NCRI)—is concerned with facilitating the aggregation of different types of data (specifically, MRI scans and histopathol-ogy slides) to aid the treatment of colorectal cancer; the second—a rather larger project funded by the
UK’s Medical Research Council (MRC) —is concerned with the development of a virtual research environment to support neuro-imaging research. In both cases, the underlying infrastructures are being developed by a team that is based in Oxford; it is the experiences of this team that we report upon in this article. We also report upon how we have considered the future potential for our systems interoperating with other systems which are deployed within the UK’s National Health Service (NHS).
introduction
The potential for distributed, service-oriented architectures to support healthcare delivery, training and research has been acknowledged widely. In this article we report upon our experiences of developing Web-services based infrastructures within two projects. In both cases, the architectures have been influenced by our earlier experiences within the e-DiaMoND project (Brady, Gavaghan, Simpson, Parada, & Highnam, 2003), which explored the development of distributed architectures to support a number of breast-cancer related applications. Although the e-DiaMoND infrastructure was based on grid services—rather than Web services—we have used many of the lessons learnt and adapted many of our designs from that project.
The two projects that we report upon are rather different in nature, but the same broad principles underpin both. The first project—a small demonstrator project funded by the UK’s National Cancer Research Institute (NCRI)—is concerned with facilitating the aggregation of different types of data to aid the treatment of colo-rectal cancer. The second project is concerned with the development of a virtual research environment to support neuro-imaging research. In both cases, the projects involve multi-disciplinary teams from a number of institutions, with the underlying infrastructures being developed by a team within Oxford. It is the experiences of this team that we report upon here.
In Power, Politou, Slaymaker, and Simpson (2005), the authors considered the information security requirements incumbent upon health grid architectures deployed within the United Kingdom, and presented an architecture for an idealised health grid that was informed by those requirements; in Power, Politou, Slaymaker, and Simpson (2006), the authors described requirements for, and an approach to, the facilitation of fine-grained access control within systems in which third party Web services are deployed.
In this article, we describe how some of the ideas from those articles have been combined to produce designs for, and implementations of, secure infrastructures, which underpin the two aforementioned e-health projects. We also comment our future intentions—which involve building on the work undertaken thus far and considering the potential for inter-operating with systems deployed within the UK’s National Health Service (NHS).
The structure of the remainder of the article is as follows. In the next section we describe the background and motivation for our work. Then we reprise the contributions of Power et al. (2005) and Power et al. (2006), which, together, provide a blueprint for our work. Next we report upon our experiences within two projects: an NCRI-funded demonstrator project and the MRC-funded Neu-roGrid project. We first introduce the projects, and then present an overview of the technologies used within our solutions. We also briefly consider some of the challenges that we have faced. Finally, we summarise the contribution of this article and outline some areas for future work, much of which is being undertaken with the GIMI (Generic Infrastructure for Medical Informatics) project (Simpson, Power, Slaymaker, & Politou, 2005).
context
A number of e-health projects have been undertaken in recent years, with the term grid computing (see, for example, Foster & Kesselman [1999] and Berman, Fox, & Hey [2003]) often being used within this context. Some interpretations of the term grid computing characterise it as the utilisation of a specific collection of services and toolkits to build a distributed architecture; other interpretations are rather looser and characterise it as the bringing together and sharing of compute and data resources from different administrative domains—in the form of a virtual organisation—to perform tasks that would otherwise be very difficult, if not impossible. In this respect, compute grids offer the opportunity to provide unparallelled processing power to facilitate, for example, analysis of 3D images, and data grids offer the opportunity to share information between sites to allow distributed data analysis.
The UK’s national e-Science Programme (Hey & Trefethen, 2002) —the main aims of which were to build a computational infrastructure to support large-scale research and to identify potential applications for such an infrastructure—funded a number of e-health projects. Such projects, including the aforementioned e-DiaMoND, have sought to develop distributed infrastructures to facilitate healthcare research, training, and delivery. Other initiatives have been seen in other countries, with examples including Singapore and Australia.
The EU HealthGrid initiative, aims (amongst other things) to promote the concept of grid computing within the biomedical community, are being undertaken to ensure that relevant technological advances developed by the grid computing community benefit healthcare research and delivery.
Simultaneously, the UK government has invested significant amounts (the initial estimate was approximately six billion pounds; the latest is approximately 12 billion pounds) in a National Programme for Information Technology (NPFIT) (since renamed Connecting for Health) in the NHS, which promises to deliver electronic records, electronic prescription of drugs and electronic booking of appointments, all of which will be underpinned by an NHS Information Technology (IT) infrastructure (Humber, 2004). Similar schemes are being developed throughout Europe and in Australia, Canada, and the United States to provide “cradle-to-grave” views of patients via the linking of electronic information (Cornwall, 2002).
While the potential benefits of the system are significant in terms of increased quality of health-care delivery, there are potential drawbacks, with many authors (see, for example, Collins [2004], Leyden [2004], Carvel [2005], Keighley [2005], and Mulholland [2005]) being critical. Critics have tended to focus on potential breaches of security and confidentiality. (The interested reader is referred to Anderson [1996] and Anderson [1999] for overviews of the relevant issues.)
It seems almost inevitable that the two paths of e-health research and systems such as Connecting for Health will converge in the near future: with real patient data stored in electronic patient records being used to support medical research. In this respect, it should be noted that research has been characterised as a “secondary use” for Connecting for Health. The following quote from the editors of the Journal of Medical Internet Research supports this view:
One aspect of electronic care records which has received little attention is the potential benefit to clinical research. Electronic records couldfacilitate new interfaces between care and research environments, leading to great improvements in the scope and efficiency of research. Benefits range from systematically generating hypotheses for research to undertaking entire studies based only on electronic record data … Clinicians and patients must have confidence in the consent, confidentiality and security arrangements for the uses of secondary data. Provided that such initiatives establish adequate information governance arrangements, within a clear ethical framework, innovative clinical research should flourish. Major benefits to patient care could ensue given sufficient development of the care-research interface via electronic records. (Powell & Buchan, 2005)
Of course, it will first be necessary to consider the confidentiality and security of patient records, and, in particular, appropriate anonymisation and pseudonymisation of data before new interfaces between healthcare delivery and clinical research environments can be facilitated. Other issues include determining appropriate consent arrangements (whether “opt-in” or “opt-out”), ensuring that trust between practitioners and patients isn’t compromised, and establishing workable governance arrangements.
Our work over the past three years has been concerned with the design, implementation, and deployment of distributed security solutions (or health grids) to facilitate medical research. Early work—such as that within e-DiaMoND—utilised Globus Toolkit 3 and enterprise-level commercial products. More recent work—for reasons of interoperability and extensibility—has been based on Web services and freely available software.
In Power et al. (2005), the authors considered the information security requirements incumbent upon health grid architectures deployed within the United Kingdom, and presented an architecture for an idealised health grid that was informed by those requirements. In Power et al. (2006), the authors described an approach to the facilitation of system-wide security that enables fine-grained access control within systems in which third party Web services are deployed. We provide a brief overview of the work of these articles in the next section before considering how the theory has been realised in practice.
Towards secure health grids
In this section, we provide brief overviews of the work of Power et al. (2005), which presented an architecture for a secure health grid, and Power et al. (2006), which described an approach to the securing of Web services.
A secure Health Grid Architecture
Our discussion is necessarily focused on the situation within the United Kingdom: other countries will have their own concerns to address. Within the United States, for example, the Health Insurance Portability and Accountability Act of 1996 (HIPAA) is of concern: the privacy rule “sets forth what uses and disclosures are authorized or required and what rights patients have with respect to their health information” (Verhanneman, Jaco, & De Win, 2003); the security rule “specifies what implementation is obligatory for enforcement of this policy or what reasonable efforts should be [undertaken]” (Verhanneman et al., 2003).
UK-based e-health projects must adhere to the principles of the Data Protection Act of 1998, which can be stated as follows.
• Personal data shall be processed fairly and lawfully (and in accordance with certain conditions).
• Personal data shall be obtained for one or more specified and lawful purposes, and shall not be further processed in any manner incompatible with that purpose or those purposes.
• Personal data shall be adequate, relevant, and not excessive in relation to the purpose or purposes for which they are processed.
• Personal data shall be accurate and, where necessary, kept up to date.
• Personal data processed for any purpose or purposes shall not be kept for longer than is necessary for that purpose or those purposes.
• Personal data shall be processed in accordance with the rights of data subjects under the Data Protection Act.
• Appropriate technical and organisational measures shall be taken against unauthorised or unlawful processing of personal data and against accidental loss or destruction of, or damage to, personal data.
• Personal data shall not be transferred to a country or territory outside the European Economic Area unless that country or territory ensures an adequate level of protection for the rights and freedoms of data subjects in relation to the processing of personal data.
The DPA is augmented by the Council of Europe’s Recommendation on the Protection of Medical Data.
If any UK-based health grid were to be deployed to facilitate healthcare delivery, or if it were to receive data from systems deployed within the NHS, then it would have to consider additional requirements. The NHS comprises a number of independent legal entities—known as hospital trusts—with each hospital trust is being responsible for the data held at its sites. This data is released only with respect to the principles of the Caldicott Guardian, which can be stated thus.
• Justify the purpose(s): every proposed use or transfer of patient-identifiable information within or from an organisation should be clearly defined and scrutinised, with continuing uses regularly reviewed by an appropriate guardian.
• Don’t use patient-identifiable information unless it is absolutely necessary.
• Use the minimum necessary patient-identifiable information.
• Access to patient-identifiable information should be on a strict need-to-know basis.
• Everyone should be aware of their responsibilities.
• Understand and comply with the law: every use of patient-identifiable information must be lawful.
It should be noted that each trust retains the ownership of all data located at its sites and each trust determines who can access its data (and under what circumstances): this is a model that we have adhered to when developing infrastructures to facilitate research. A goal of our work, then, is to design and deploy systems that allow data to be shared as ensuring that the data owner retains absolute control over who can access which data, when it can be accessed, and even where it can be accessed from.
To this end, a number of use cases—pertaining to information security requirements—were presented in Power et al. (2005). We provide an overview of these below.
• Distributed queries of patient data. “A user wishes to query the data held on a subset of the hospitals that form the health grid. Each hospital is allowed to decide its own policy for data access. The user should receive the combined results containing only data that they are permitted to access.”
• Working at a remote site. “A doctor is working at a remote hospital, which is part of the health grid. The doctor should be able to access data from their home hospital, though their request may be subject to a policy that differs from the one used when they are at their home institution.”
• Delegation of access permissions. “A senior health professional would like to grant access to data to a colleague. This access should be temporary, and could be granted to either a named individual or a group of people.”
• External access. “Either a health professional working from home or an individual patient wishing to see their own records should be able to access data in accordance with the local hospital’s access control policy. The hospital would use a different policy for such external access than would be used for requests from a remote hospital. This use case differs from the others as the request comes from outside of the current virtual organisation.”
• Modification of data. “Having made a clinical decision about a case, a doctor wishes to modify the data stored in the health grid. A doctor will only be able to modify data if a hospital’s policy allows it. Each hospital is responsible for the data it stores and as such it should keep a record of all modifications made. This use case is similar to the delegation of access permissions described above, with the only difference being that the data—rather than the policy—is being changed.”
• Transferring patient records. “In this use case a patient has moved and is now being treated at a new hospital. As the patient is likely to stay at the new hospital for some time, it would make sense to move their data. To be able to move the data it will first need to be read: this may involve a distributed query as data may already be present at other hospitals. The data will then need to be deleted from one hospital and copied to another—as the responsibility for it has transferred. This will involve the modification of data. Finally the access policies at both of the hospitals may need to be changed to reflect the change of ownership of the data.”
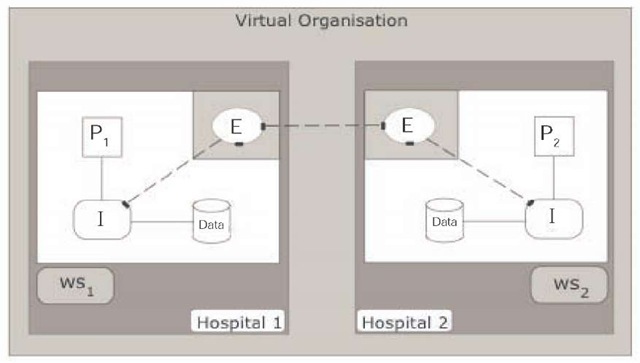
In the architecture of Power et al. (2005), each node contains a data store, externally facing services, internally facing services, access control policies, and workstations (see Figure 1). (This situation is visualised in terms of a health grid with two nodes above.) It is the externally facing services that allow different sites to communicate with each other. Importantly, this architecture allows each site to retain control of its data and access control policies. All access requests within this system are governed by the policies accessible to the internal service, with the policies governing who can access the data, when they can access it, where it can be accessed from, and what rights they have to delegate that access.
In this architecture, a local user can make a request to its local externally facing service, which would then direct that request to: the local internal service; another external service (or set of external services) at a remote site; or both the local internal service and other sites’ external services.
securing Web services
In Power et al. (2006) an approach to the facilitation of system-wide security that enables fine-grained access control within systems in which third party Web services are deployed was described; in particular, a characterisation of security features required to enable existing Web services to be secured to fit in with a secure infrastructure was presented. We provide an overview of some of these requirements here.
Figure 1.
• Authenticating clients. Our assumption is that the system of interest already has an established authentication mechanism. The existing Web service, however, may utilise a different authentication mechanism. When writing a wrapper service, it is possible to authenticate clients using the system-wide authentication mechanism and then translate the system-wide client identifier into one that is understood by the existing Web service via the utilisation of a client mapping function. Once the mapping has been performed, the original SOAP message can be passed to the existing service using whatever authentication mechanism it supports.
• Secure messaging. If the content of the message is encrypted, then the message will need to be decrypted and possibly encrypted again. Furthermore, the keys used between the client and the wrapper will differ from those used between the wrapper and the Web service.
• Access control. The resource which we are trying to provide access to will typically have its own access control mechanism. Ideally, to be consistent with the requirements of Section 3.1, all access control for the resources at a single node should be determined by a single set of coordinated policies, with all requests for access to the existing Web service having to comply with these policies.
A PLATFORM FOR DATA AGGREGATION TO SUPPORT
colorectal cancer applications
The National Cancer Research Institute (NCRI) Informatics Initiative has facilitated funding for a nine-month demonstrator project to bring together researchers from various disciplines to demonstrate the utility of a multi-scale, multi-disciplinary approach to enhancing the information that can be derived from data collected within a clinical trial. The intention is to develop a prototype system to relate MRI images to the consequence macro images, and to demonstrate the application of medical image analysis (developed for radiological scale images) to macro slides.
The development of a secure computing infrastructure to link disparate sites is being undertaken at Oxford University Computing Laboratory; the development of a prototype viewing application is the responsibility of the Department of Engineering Science at Oxford; the development of appropriate ontologies and meta-models is being undertaking by the Department of Computer Science at University College, London; and data collection is being undertaken by the Pathology Department at Leeds University and the Royal Marsden Hospital. Finally, the task of coordinating all of this activity is the responsibility of the NCRI Informatics Coordination Unit.
In Slaymaker et al., 2006), the development of the underlying computing infrastructure was reported. In this section, we characterise this development in terms of the requirements of a previous section.
Colorectal cancer is first diagnosed using endoscopy and confirmed by histopathology. It is then staged radiologically in order to determine the extent of local and distant disease—with this being best done using MRI. The primary tumour site is assessed to determine if the tumour has extended into the adjacent fat and involved the adjacent tissue planes, including the mesorectum; the detection of lymph nodes and their possible infiltration by the tumour is also crucial, with distant metastases being assessed separately using Computed Tomography (CT) or (if appropriate) Positron Emission Tomography (PET).
Typically, an oncologist sends a patient who is suspected of suffering from colorectal cancer to a radiologist for a CT scan: suspicious dense areas that could be cancer may be revealed; the scans may also give early evidence that, in the case of colorectal cancer, there is already strong evidence of metastasis. A number of options are available on the basis of the CT examination: “palliative care”; sending the patient for MRI scans for further information; providing a course of chemo- or radiotherapy; or surgery. The project is concerned with the last of these options.
During surgery, the surgeon cuts out the tumour. The extracted tumour, together with some flesh, is photographed from the front and the back. The tumour is divided into slices of three-mm thickness, with each slice being captured in slides with two different resolutions: a low-resolution image is taken at about x20 zoom (macroscopic resolution), and a high resolution is taken at about x140 zoom (microscopic resolution). The microscopic slides are analysed by histopathologists to assess type and stage of a cancer, with this analysis consisting of considering the distribution, shape variation, and staining of cells visible at the higher resolution.
The Royal Marsden Hospital has collected (anonymised) MRI volumes of colorectal tumours, with all MRI volumes being taken prior to chemotherapy/radiotherapy and prior to surgery. These volumes are also accompanied with descriptive metadata such as the MRI position, the extent of the tumour, the surgical plan, etc. The other data resource that the project draws upon is a collection of macroscopic and microscopic slides in digital form supplied by Leeds University, with each micro image being stored in a custom file format.
Neurogrid
Currently, neuro-imaging research typically consists of small studies being carried out in single centres. The sharing of data between centres is limited and even when data is shared, it is common for researchers to be very guarded with their data and algorithms—which leads to much duplication of effort.
The NeuroGrid project (Geddes et al., 2005, Geddes et al., 2006), which is funded by the UK’s Medical Research Council, is concerned with tackling problems that are currently holding back widespread data sharing, with the principal aim of the three-year project being to develop a distributed collaborative research environment to support the work of neuroscientists. The potential benefits of the NeuroGrid platform include the streamlining of data acquisition, the aiding of data analysis, and providing improvements to the power and applicability of studies. The project involves collaborators from the University of Oxford, University and Imperial Colleges, London, Nottingham University, Edinburgh University, Cambridge University, and Newcastle University.
As a means of ensuring that the technological solutions being developed within NeuroGrid are of clinical relevance, the project is focusing on a number of exemplars, which pertain to Dementia, Stroke, and Psychosis. The Dementia exemplar requires real-time transfer and processing of images with a view to assuring image quality prior to the patient leaving the examination. The Stroke exemplar is establishing and testing mechanisms for interpretation and curation of image data which are essential to the infrastructure of many multi-centre trials in common brain disorders. The Psychosis exemplar is testing the capabilities of NeuroGrid to: deal with retrospective data, assimilate material into databases, and facilitate data analysis.
solutions
In this section we describe the solutions deployed within the projects described previously. Although the approaches taken in both cases are broadly similar we consider different aspects in our discussions of both projects to reflect the different natures of the two projects: first we concentrate on database aspects; and then we concentrate more on Web services.
An Infrastructure for Data Aggregation to support colorectal cancer Applications
It is essential that it is verified that both MRI and pathology images come from the same patient cases, and that these two sets of data can be related (both technically and ethically). If this is the case, we can then relate and integrate pathology and radiology images of a case in two ways. The first means of relating these different types of data is via the integration of the full images as entities—which means having available the pathology image one can retrieve together with its associated MRI image/report. The second method is via the association of their corresponding region of interests (ROI) —which means if one has a ROI in an MRI volume, then one should be able to retrieve its associated area in the 3D pathology images.
Typically, each MRI volume can have more than one ROI, because individual lymph nodes could also be annotated as well as the main tumour (although only one of these annotations would apply to a lesion). Thus, a flexible database schema that can effectively describe all of the above data is necessary. The database underpinning the project is based on the work described in Power, Politou, Slaymaker, Harris, and Simpson (2004), in which a relational structure for images stored in the standard Digital Imaging and Communications in Medicine (DICOM) format is presented.
As the pathology images are not in DICOM format, a different method of dealing with them is required. The two formats that need to be handled are JPG for the macroscopic images and a custom file format for the microscopic images. We have chosen to employ a method that effectively automatically generates suitable DICOM wrappers for each image file. This has the benefit of allowing maximum flexibility while requiring minimal changes to the existing underlying schema.
Another advantage of using DICOM-style wrapping is that it handles collections of images well. A single set of macro images would form a DICOM series and hence be addressed together. This is also the case with the micro images. Furthermore, the two series—macro and micro—can be collected together as a single study. This provides a great deal of flexibility when processing queries.
All data is stored in a federated database that is distributed over several sites, with all of the data servers being exposed via Web service interfaces. The patient and image data are stored at the site at which they originated, with the data from each location reflecting the specialisation of that site. The selection of MRI and pathology data pertaining to a single case is achieved by federating data between multiple sites. The architecture is based on that of a prior section and utilises open source Web services technologies and standards.
The NeuroGrid Infrastructure
While our developments within NeuroGrid have followed the sentiments of the architecture of Power et al. (2005), the system is by no means a direct implementation. For example, there has been no need to distinguish between internal and external services as both kinds of service have been written and deployed by the NeuroGrid core technology team. Should there be any need in the future to decouple the internal and external services (if, for example, a third party internal service were to be used), then this could be achieved via the existing services calling through to the internal services.
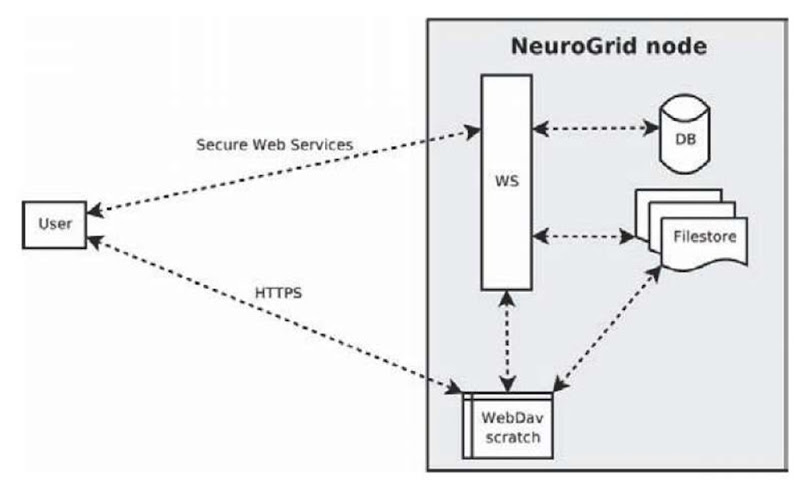
The configuration of a NeuroGrid server node is illustrated in Figure 2.
Ensuring secure messaging was our first challenge, and we use secure Web services to this end. In addition, we utilise WebDAV (Web-based Distributed Authoring and Versioning) (Whitehead, 1998) folders to provide scratch space for operations. The Web services and WebDAV folders are accessible only by using PKI mutual authentication using X.509 certificates. The Web services use an insecure channel to carry secure messages signed by the originator and encrypted for the recipient, ensuring both the identity of the sender and secrecy. The users communicate with the WebDAV server over HTTPS with mutual authentication enabled—which establishes a secure channel over which insecure messages can be passed.
Figure 2.
WebDAV plays a key role in the NeuroGrid architecture. First, there is no standard high-speed and secure way of transferring binary data using SOAP messages; WebDAV allows us to both get files from and put files to Web servers in a secure way efficiently. Second, WebDAV allows the user to browse, upload and download files directly to their scratch space using a variety of third party WebDAV clients—this is especially useful when wrapping the Web services with a portal. The WebDAV folders also provide an ideal location for any intermediate results that might be generated by algorithms which do not belong in the file-store but may be of interest to the user.
A file-store and a database of meta-data provide part of the back-end of the system: this is where data is stored, with all interactions occurring through Web service calls. We separate image data from patient and meta-data (with appropriate references between the two to guarantee referential integrity). The use of the Apache Derby data base—which is written entirely in Java, is easy to install, and has a minimal system footprint—was chosen as it enhances the interoperability and portability of the underlying infrastructure.
We are also using Sun’s JWSDP (Java Web Services Development Pack) Web service implementation in the Sun customised version of the Apache Tomcat container. The intention behind using Java-based technologies is that it should allow us to deploy to any platform with a Java Virtual Machine; using Web services allows maximum interoperability with client implementations in a variety of languages. We are also using the Apache Web server which is available for most platforms.
The technologies we have used to create the NeuroGrid infrastructure were chosen to ensure that it would be as portable and interoperable as possible: we have attempted to choose solutions with excellent cross-platform support and which is at freely available—if not open source.
We now consider how the use cases of the third section are being realised.
• Distributed queries of patient data. This use case requires the ability to perform federated queries—an ability that the NeuroGrid infrastructure provides. To enable federated queries the system must support onward Web service calls—a situation in which one Web service makes a call to another Web service, which can be local or on a remote node. A request to a federated query Web service at one site will query local resources and, in addition, make Web service query calls to other known sites. On receipt of the results of its Web service queries the federated Web service assembles all the data into a single set of results and returns this to the caller of the federated Web service.
In order to identify the originator of the request, a ticketing system is being used. The ticketing system iteratively propagates information pertaining to authentication: each Web service call has a ticket associated with it and each Web service examines the tickets it receives. When a Web service creates a new ticket, the calling Web service’s ticket (or a ticket issued to the calling user in the initial case) is embedded in the new ticket. Tickets are always signed by the sender ensuring the validity of the ticket can be verified by all subsequent receivers. The tickets constitute a verifiable audit trail which can be used to restrict access control when combined with a relevant access control policy.
• Working at a remote hospital. This simple use case again relies on the ticket system. At the points when access control policies are evaluated, the audit trail of the ticket can be consulted and that shows exactly the route the user has taken to make the request. Access can then be restricted by combining a policy which restricts access based on origin and/or route.
NeuroGrid already has the ability to onwardly call Web services (one Web service making a call to another Web service). Using this capability a user can make calls to a node at the hospital where they are currently located and that node will make appropriate requests to the user’s home hospital on their behalf.
In a hospital situation it would be appropriate to have nodes which could only be communicated with locally—apart from node-to-node communication which is essential to the system’s operation. This could be achieved by several means, such as IP address restrictions or the utilisation of a Virtual Private Network.
In the currently deployed NeuroGrid system, nodes are all public facing, which means that a user could bypass the step of calling the Web services on a local node and instead call directly to a remote node.
• Delegation of access permissions. In the original presentation of the use case of Power et al. (2005), the delegation of access permissions was similar to that pertaining to the modification of data; within NeuroGrid we are also looking at a model of delegation based on the aforementioned ticket system.
Using the ticket system a user can delegate their permissions to access data to another user. Using the ticket system, the originator of the request is known—and thus their identity can be used in making access control decisions. This information can also be used to delegate authority from one user to another.
• External access. In Power et al. (2005), external access referred to any access that was not initiated from within a site of the virtual organisation, such as a doctor accessing some medical records from home. From the perspective of the NeuroGrid infrastructure, all access is considered external: in the current system, physical location does not affect access rights.
There are two ways to access the NeuroGrid system: talking directly to a server using an application that utilises an API or through a Web portal (which itself is just an application using the API).
It is trivial to establish external access as any location not in a restricted list. This could then be used to deny direct access to NeuroGrid to any location not on the list. Locations not on the list could access the NeuroGrid through the Web portal—with more restricted rights than permitted to an internal user.
It is well understood that it is difficult to establish a definite location of a client on a network. As such, it could be argued that this “all or nothing” (without going through the portal) model represents a sensible approach: the users’ rights would simply be restricted when accessing the system though a portal. Additionally, there is no reason why externally-facing NeuroGrid nodes could not be configured to give the same reduced rights to users directly through Web services.
The access control policies can take advantage of the tickets to accomplish this restricted access as they contain the audit trail of the request: any requests which have passed through a specific node can be given restricted access rights.
• Modification of data. Within the current implementation, it is perfectly possible to modify the data held in the system; all modifications are recorded as part of an audit system, so changes to the data can be tracked back to the responsible party if necessary. Again, the implementation of this use case is benefiting from the ticket system (combined with access control policies) to allow and disallow rights to modify data as necessary.
• Transferring patient records. It is entirely possible within NeuroGrid to copy some data (in this case, a patient record) to another node. Ideally, this process would take place within the scope of an atomic transaction ensuring that the patient record is neither duplicated, nor, more seriously, lost. Support for atomic transactions is part of the WS-Transaction specification, and support for this specification is under development as part of a future version of JWSDP. Once this becomes available we plan to add trans-actional support to the NeuroGrid system.
As any deployment of the NeuroGrid infrastructure evolves, modifications of access control policies—to reflect the change in location of data—will occur. For a standard record, this will typically mean that it is added to a list of records associated with a standard access control policy at the hospital it was transferred to—with any relevant specific policies for that patient also being transferred.
challenges
Our goal is to develop secure distributed systems that will support a wide range of platforms: by using open standards we aim to support interaction with third party systems that may be developed in the future. By using a platform-independent programming language (Java) and open standards for communication (HTTP, SOAP) and security (TLS, WS-Security) we should—in theory—be able to meet these objectives. We also aim for our systems to be free to deploy and use—which prohibits the use of most commercially produced software.
Although large vendors provide free versions of commercially available software, they (typically) provide limited support for its use. In spite of this, we have chosen to use several software packages that fall into this category alongside some open source components. We discuss two of the challenges that we have faced in this regard in the following.
The distributed systems we have developed consist of multiple components, which communicate using a combination of WebDAV and Web services. The WebDAV services are provided using an Apache Httpd server with DAV and SSL extensions—which allows us to provide a mutually authenticated WebDAV service. The URLs used tend to be long—for technical and security reasons (and user convenience). This had led to several problems on the client-side, where ideally we would like to use the default WebDAV clients provided as part of the operating system.
Microsoft provides a WebDAV client built into Internet Explorer and also support the opening of WebDAV folders as a Network Connection. We have had two problems with these clients. The first is the lack of support for long URLs in Internet Explorer—a series of support calls over a period of several months led to a suggestion that we shorten the URLs. We also have had problems with Microsoft clients trying to write to the root of the server’s file system as this would be normal behaviour on a Microsoft server. Apple also provides a WebDAV client which is used primarily for their Mac services. This client has the problem that it does not support mutual authentication.
As a means of providing Web services, we use the Sun Java Web Services Development Pack (JWSDP) running on top of a Sun provided version of Tomcat 5.0 (Sun do not currently support a version of Tomcat 5.5 although the reason for this is unclear). Part of our reason for choosing the Sun provided software instead of the open source Axis was because of its support for a large number of the Web service standards. Unfortunately, the WS-security libraries require extensions to the Java security libraries—which are shipped with all Sun-provided JVMs, but not IBM-provided JVMs. This has led to problems running our Web services on IBM servers.
There are a number of lessons to be learnt from this. First, the use of Web services to develop real, interoperable systems to support genuine virtual organisations still has some way to go. Second, it is arguable that the use of extensible standards to facilitate interoperability is doomed to failure as long as vendors are able to release product versions underpinned by their own closed-source extensions to these open standards.
Discussion
In this article we have described how some of the requirements and approaches described in Power et al. (2005) and Power et al. (2006)—pertaining to the design of health grid architectures and the securing of Web services, respectively—have been realised in two e-health projects.
Developing robust distributed solutions using current technologies is a challenging task. One of the major issues that we have faced is that current toolkits for Web services are very much focused on a single server being contacted by multiple clients; as such, are merely a substitute for a traditional HTML-based solution. We would argue that inadequate thought has been given to building multiple-server applications. For example, we have encountered several problems when making onward calls from one Web service to another. Other challenges include “standard” extensions not being standard and a lack of a mechanism for transferring large binary objects that is both secure and efficient simultaneously.
Future work is being taken in a number of directions, much of which is being undertaken within the GIMI (Generic Infrastructure for Medical Informatics) project (Simpson et al., 2005). The main aim of GIMI is to develop a generic, dependable middleware layer capable of: (in the medium-term) supporting secure and ethical data sharing across disparate sources to facilitate healthcare research, delivery, and training; and (in the longer-term) interfacing with technological solutions deployed within the NHS.
The key to ensuring legal and ethical access to such data is that the mechanism should offer fine-grained and dynamic access control to resources: this is the key driver behind GIMI. The infrastructure will be developed so that it is sympathetic to the needs of clinical researchers, commercial organisations involved in the medical domain, healthcare providers, and those concerned with providing training facilities—all of which will be concerned with sharing confidential data. Our plan is to extend our existing designs and implementations to facilitate such means of authorisation.
First, we have made significant strides towards realising the architecture of Power et al. (2005). There is some way to go, however: the addressing of the “transferring of patient records” use case, for example, has yet to be addressed. Continuing to realise these use cases within the context of the NeuroGrid project shall continue to be a priority.
Second, our architectures have been designed with the opportunity for fine-grained and flexible access control policies in mind. Our medium-term goal is to realise this opportunity by utilising XACML to secure nodes in such a fashion. This—together with the long-term interoperability view—is our focus within GIMI.
Finally, to date, our techniques have been deployed exclusively within the healthcare domain; we intend, in the near future, to determine other relevant application domains in which our approaches may be validated.