Systematics is the study of biological diversity that has as its , primary goal the reconstruction of phylogeny. the evolutionary or genealogical history of a particular group of organisms (e.g., species). Because of its emphasis on phylogeny. this discipline is often referred to as phylogenetic systematics or cladistics. Other related goals of systematics include determination of the times at which species originated and became extinct and the origin and rate of change in their characteristics. An important component of systematics is taxonomy, the identification, description, nomenclature, and classification of organisms. Systematics provides a framework for interpreting patterns and processes in evolution using explicit, testable hypotheses.
The rapid pace of research on marine mammals has resulted in renewed interest in their systematics. Phylogenetic systematic methodology as introduced here has gained near universal acceptance. [For a general introduction to the topic readers are referred to texts by Eldredge and Cracraft (1980), Wiley (1980), and Wiley et al (1991).] In addition to their use in elucidating evolutionary relationships, phylogenies are now recognized as powerful tools for unveiling evolutionary patterns of marine mammal diversity in ecological and behavioral settings.
I. Basic Tenets of Phylogenetic Systematics
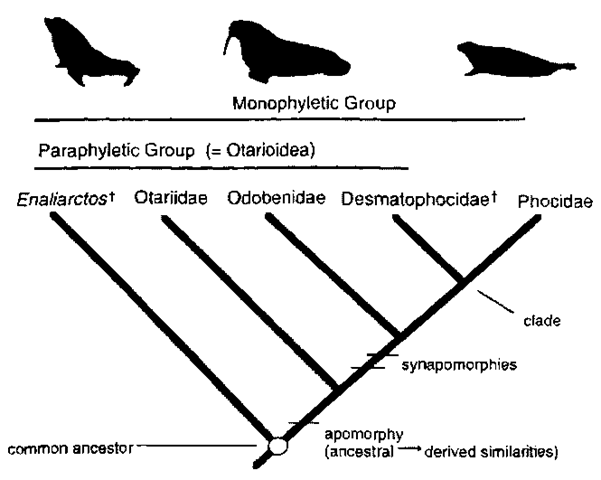
The recognition of patterns of relationship among species is founded on the concept of evolution. Patterns of relationship among species are based on changes in the characters of an organism. Characters are diverse, heritable attributes of organisms that include DNA base pairs, anatomical and physiological features, and behavioral traits. Two or more forms of a given character are termed character states. For example, among pinnipeds, the character contact between maxillary and frontal bones consists of three character states: (1) V shaped (in bears, extinct desmatophocids, Enaliarctos, and phocids). (2) W shaped (in otariids), and (3) transverse (walruses) as described by Berta and Sumich (1999). In the establishment of relationships among groups of organisms, phylogenetic systematics emphasizes evolutionary novelties (derived characters) in contrast to ancestral similarities (primitive characters).
The evolutionary history of a group of organisms can be inferred by sequentially linking species together based on their common possession of derived characters, also known as synapo-morphies. If derived characters are unique to a particular taxon rather than showing relationships among taxa they are termed autapomorphies. An example of an autapomorphy is the transverse contact between maxillary and frontal bones seen only in walruses among pinnipedimorphs (living pinnipeds and their fossil relatives). Derived characters are considered to be homologous, a similarity that results from common ancestry. For example, the flipper of a seal and that of a walrus are homologous because their common ancestor had flippers. In contrast to homology, a similarity not due to homology is honwplasy. For example, the flipper of a pinniped and that of a whale are homo-plasious as flippers because their common ancestor lacked flippers. Homoplasy may arise in one of two ways: convergence (parallelism) or reversal. Convergence is the independent evolution of a similar feature in two or more lineages. Thus seal flippers and whale flippers evolved independently as swimming appendages: their similarity is homoplasious by convergent evolution. Reversal is the loss of a derived feature coupled with the reestablishment of an ancestral feature. For example, in phocine seals (bearded seal Erignathus barbatus, hooded seal Cystophora cristata. and the Phocini) the development of strong claws, lengthening of the third digit of the foot, and deemphasis of the first digit of the hand are character reversals because none of them characterize phocids ancestrally but are present in terrestrial arctoid carnivores, common ancestors of pinnipeds (Berta and Sumich. 1999).
Relationships among organism groups are commonly represented in the form of a cladogram, a branching diagram that conceptually represents the best estimate of phylogeny (Fig. 1). Derived characters are used to link monophyletic groups, groups of taxa that consist of a common ancestor plus all descendants of that ancestor (referred to as a clade). For example, a hypothesis of relationships among pinnipedimorphs (inclusive group that includes all pinnipeds and their close fossil relatives) based on morphologic characters proposes that phocid seals (Phocidae) and an extinct lineage (Desmatophocidae) are more closely related to each other than either is to walruses (Odobenidae). Fur seals and sea lions (Otariidae) are positioned as the next closest relative to this clade (walruses + desmatophocids + phocids) with the fossil taxon Enaliarctos recognized as the most basal lineage (Berta and Sumich, 1999; Fig. 1). According to this hypothesis, relationships among pinnipedimorphs are depicted as sets of nested hierarchies. In this case, four monophyletic groups can be recognized. The most exclusive monophyletic group is that formed by phocid seals and desmatophocids, as this clade shares derived synapomor-phies not also exhibited by walruses, otariids. or Enaliarctos. At the other extreme, the most inclusive monophyletic group is that formed by Enaliarctos + Otariidae + Odobenidae + Des-matophocidae + Phocidae.
Figure 1 Hypothesis of pinnipedimorph relationships, f, extinct taxa.
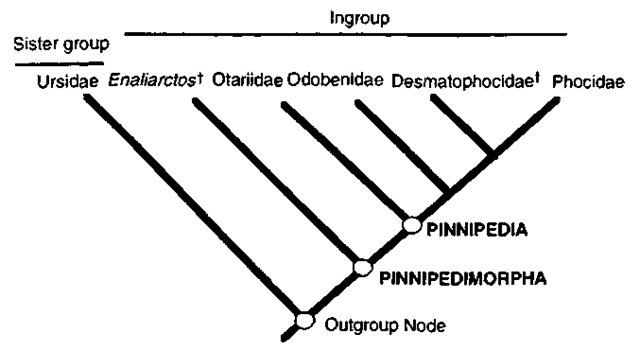
The task in inferring a phylogeny for a group of organisms is to determine which characters are derived and which are ancestral. If the ancestral condition of a character or character state is established, then the direction of evolution from ancestral to derived can be inferred, and synapomorphies can be recognized. The methodology for inferring the direction of character evolution is critical to cladistic analysis. Outgroup comparison is the most widely used procedure. It relies on the argument that a character state found in close relatives of a group (the outgroup) is likely to be the ancestral or primitive state for the group of organisms in question (the ingroup). Usually more than one outgroup is used in an analysis, the most important being the first or genealogically closest outgroup to the ingroup called the sister group (Maddison et al. 1984). For example, among pinnipedimorphs the ingroup includes the Phocidae (seals), the Desmatophocidae (extinct seal relatives), the Otariidae (fur seals and sea lions), the Odobenidae (walruses) and the fossil taxon Enaliarctos. The Phocidae is hypothesized as the sister group of the Desmatophocidae and these taxa together form the sister taxon to the Odobenidae. The Otariidae and Enaliarctos are positioned as sequential sister taxa. In the hypothesis presented here, bears (Ursidae) are positioned as the closest pinniped outgroup (Fig. 2), although alternate hypotheses support pinnipeds as either allied with mustelids or as having an unresolved ancestry with arctoids (ursids, mustelids, and procyonids).
Figure 2 Pinniped relationships with ingroup and outgroups identified, f, extinct taxa.
II. Phylogeny Reconstruction
The first step in the reconstruction of phylogeny of a group of organisms is selection and definition of characters and character states for each taxon (e.g., species). Next, the characters and their states are arranged in a data matrix (see Table I). Characters can be further distinguished; those with two states are binary, whereas characters with three or more states are multistate. For each character, ancestral and derived states are determined. The determination of character state, whether ancestral or derived (also called polarity assessment), is done using outgroup comparison. For example, if the distribution of character No. 1, condition of the lacrimal bone, is considered, two character states are recognized: presence versus absence (or fusion of the lacrimal such that it does not contact the ju-gal). The outgroup (bears) possess a lacrimal bone condition, which is the ancestral state (Table 1). Ingroup taxa lack the lacrimal bone, which is the derived condition. Because this derived state unites pinnipedimorphs to the exclusion of bears, it is considered a synapomorphy.
TABLE 1
Data Set for Analysis of Pinnipedimorphs Plus an Outgroup Showing Five Characters and Their Character States
|
|
|
Character/character states |
|
|
|
|
|
1 |
2 |
3 |
4 |
5 |
|
Taxon |
Lacrimal bone |
Middle ear bones |
Orbital/maxilla |
Maxilla/frontal |
Squamosal/jugal |
|
Outgroup |
|
|
|
|
|
|
Ursids |
Present |
Small |
No |
V shape |
Overlapping |
|
Ingroup |
|
|
|
|
|
|
Enaliarctos |
Absent |
Small |
No |
V shape |
Overlapping |
|
Otariidae |
Absent |
Small |
Yes |
W shape |
Overlapping |
|
Desmatophocidae |
Absent |
Large |
Yes |
V shape |
Interlocking |
|
Phocidae |
Absent |
Large |
Yes |
V shape |
Interlocking |
|
Odobenidae |
Absent |
Large |
Yes |
Straight |
Overlapping |
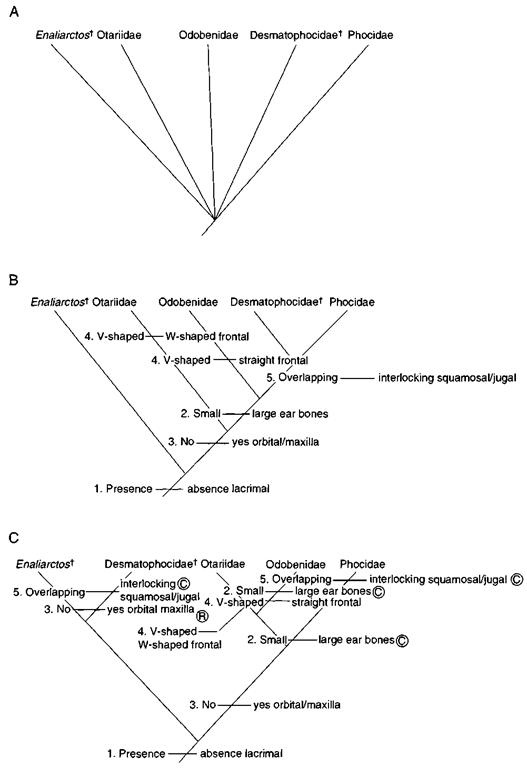
Figure 3 Three possible eladograms (A-C) of relationships and character-state distributions for the in groups listed in Table I. Convergences and reversal are denoted by a circled C and R, respectively.
A final step in phylogeny reconstruction is the construction of all possible eladograms by sequentially grouping taxa based on the common possession of one or more shared derived character states (Fig. 3). An important aspect of phylogeny reconstruction is the principle of parsimony. The basic tenet of parsimony is that the cladogram that contains the fewest number of evolutionary steps, or changes between character states of a given character summed for all characters, is accepted as being the best estimate of phylogeny. In this example. Fig. 3B is the most parsimonious cladogram. Note that an alternative cladogram for the data set (Fig. 3C) showing different relationships among the five taxa requires nine characters state changes, three more than the most parsimonious cladogram (Fig. 3B).
The methods used to search for the most parsimonious tree depend on the size and complexity of the data matrix. These methods are available in several computer programs [e.g., PAUP (Swofford. 1998) and MacClade (Maddison and Mad-dison, 1992)]. The latter is particularly useful in assessing the evolution of characters. Systematists have become concerned about the relative accuracy of phylogenetic trees (i.e., how much confidence can be placed in a particular phylogenetic reconstruction). Various measures (e.g., tree length) and indices (e.g., consistency index, rescaled consistency index) have been devised to address this concern (reviewed bv Swofford, 1998). Another approach that is often used to estimate the value of a particular cladogram with respect to data places confidence limits on the individual branches, a technique known as bootstrapping. Another concern stems from the realization that an increase in the size of data sets results in a greater chance of the analysis resulting in more than one equally parsimonious tree. A method of working with multiple trees involves the implementation of consensti.s trees that are useful in identifying the areas of agreement and conflict among competing trees. Perhaps the greatest influence on the accuracy of a cladogram is the number of taxa. When only a few taxa are used (e.g., exemplars of a group), or when only a small number are available, the accuracy of the phylogenetic reconstruction suffers. In summary, many factors should be considered in evaluating phylogenetic hypotheses, among the most important being taxonomic sampling and rigorous analysis, including the underlying assumptions of various methods.
III. Phylogenetic Classification
Taxonomy is the language of biology. One aspect of taxonomy is the classification of organisms that allows us to organize and communicate information about life’s diversity. Phylogenetic systematists contend that classification should be based on phylogeny and should include only monophyletic groups. In contrast to monophyletic groups, a paraphyletic group (designated by quotation marks) is one that includes a common ancestor and some but not all of the descendants of that ancestor. An example of a paraphyletic taxon is the “Otarioidea,” a group that includes walruses, otariid seals, and their extinct relatives to the exclusion of phocid seals (see Fig. 1). The recognition of paraphyletic taxa is to be avoided since by doing so we risk misinterpreting the evolutionary relationships of taxa and their classification. One result of the use of phylogenetic methodology is that traditionally accepted ranks (e.g., phylum, class, order, family, genus, and species) often do not correspond to new information about evolutionary relationships among taxa, thus rendering their classification misleading. One method is the elimination of rank altogether and the indication of relative rank by subordination as shown by indentation. An example, the classification of major lineages of pinnipedimorphs, is shown in Fig. 4 based on the phylogeny in Fig. 2.
IV. Uses of a Phylogeny
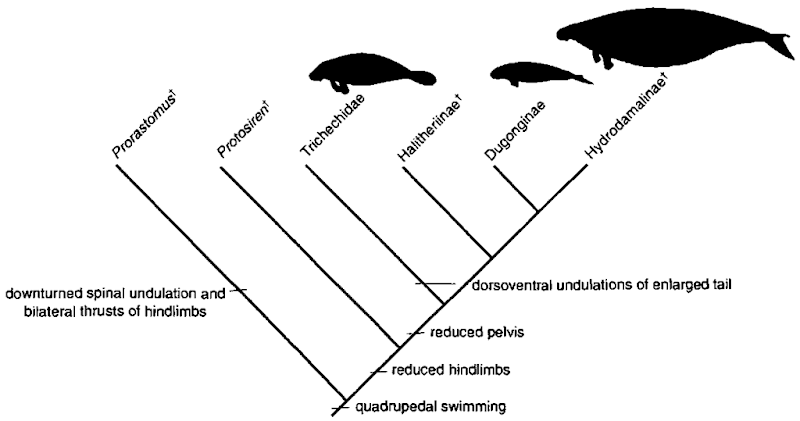
Once a phylogenetic framework is produced, one of its most interesting uses is to elucidate questions that integrate evolution, behavior, and ecology. One technique used to facilitate such evolutionary studies is optimization or mapping. Once a cladogram is constructed, a feature or condition is selected to be examined in light of the phylogeny of the group. Examples using marine mammals include the evolution of body size in pinnipeds (Wyss, 1994), pinniped host-parasite associations (Hoberg, 1995), and the evolution of locomotor and feeding behavior in pinnipeds (Berta amd Adam, 2001), cetaceans and sirenians (Domning, 2000). One of these studies, the evolution of locomotion among sirenians, is briefly reviewed here. A cladogram of relationships among sirenians was first established based on morphologic data. Next, using this phylogenetic framework, Domning (2000; Fig. 5) mapped locomotor characters onto the tree. Sirenians passed through three locomotor stages from a terrestrial quadrupedal ancestry. In the first stage exemplified by archaic sirenians, the prorastomids, swimming was accomplished by alternate thrusts of the hindlimbs. This was followed by a second stage seen in the extinct taxon Protosiren that employed dorsoventral spinal undulation and bilateral thrusts of the hindlimb in swimming. In a final stage, seen in modern sea cows and manatees, sirenians have evolved into completely aquatic animals, swimming with the tail only.
Pinnipedimorpha (including Enaliarctos and all other pinnipeds) Pinnipedia (fur seals, sea lions, seals, walruses and their extinct relatives) Otariidae (fur seals and sea lions) Phocomorpha (walruses, seals, and their extinct relatives) Phocoidea (including Allodesmus, Desmatophoca, seals and walruses) Phocidae (seals) Odobenidae (walruses)
Figure 4 Classification of the Pinnipedimorpha.
Figure 5 Evolution of locomotion among sirenians. t, extinct taxa. Based on Domning (2000).