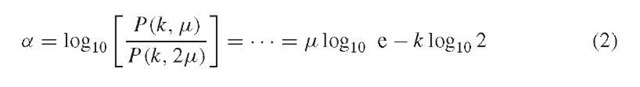1. Introduction
Historians will undoubtedly rank the Human Genome Project (HGP) among the great achievements of humanity. HGP (see Article 24, The Human Genome Project, Volume 3) represented an advancement of more than an order of magnitude in size over preliminary “model organism” projects for Caenorhabditis elegans (C. elegans Sequencing Consortium, 1998) and Drosophila melanogaster (Adams et al., 2000), which themselves were breakthroughs. A notable aspect of HGP was the problem of assembling data to deduce the original sequence. In this article, we briefly outline the concept of assembly and the complications arising from various factors, especially sequence repeats. We then describe the two different approaches applied by investigators to generate and assemble human genome data. Finally, we outline the still-evolving assessment of these two projects and attempt to place them in a historic context.
2. The concepts of shotgun sequencing and assembly
In any project, the length of DNA that can be resolved by a single sequencing reaction is invariably a small fraction of the source DNA whose sequence is desired. At their most fundamental level, projects must therefore take the so-called shotgun approach (Sanger et al., 1980; Anderson, 1981; Deininger, 1983), whereby small random fragments are read and then reassembled to deduce the original sequence. The assembly process is enabled by oversampling the genome, so that many overlapping substrings of DNA sequence are available for reconstruction.
In a purely random sequence, the assembly problem would be relatively straightforward. The population of repeated sequence strings of a given size decreases in a power-law fashion. In other words, given a specific DNA fragment of length L, a genome would need to be about 4L bases in size before we would expect to find another copy of this sequence. Disregarding sequencing errors for the moment, suppose we find two human reads that share a specific 30-bp sequence. In random sequence, such a segment would only be expected about every 430 « 1.2 x 1018 bases. Because the likelihood of finding two copies in the comparatively paltry 3 x 109 human genome would then be exceedingly small, we would be quite confident in assembling our two reads into a sequence contig. Continuation of this process would result in a gradual conglomeration of reads into ever-larger contiguous regions. In the limit, the complete sequence would eventually be recovered.
Unfortunately, actual DNA contains significant biases, including repeat structures that occur in many largely identical copies. Many instances are much longer than the length of a single sequence read, for example, LINE elements and segmental duplications (Samonte and Eichler, 2002). Sequence data derived from such regions may not be able to be uniquely, and thus, correctly placed in contigs. Consequently, there will be many admissible assemblies, only one of which is correct (Myers, 1999). This phenomenon was especially problematic in the HGP because the human genome contains a large and diverse population of repeat structures.
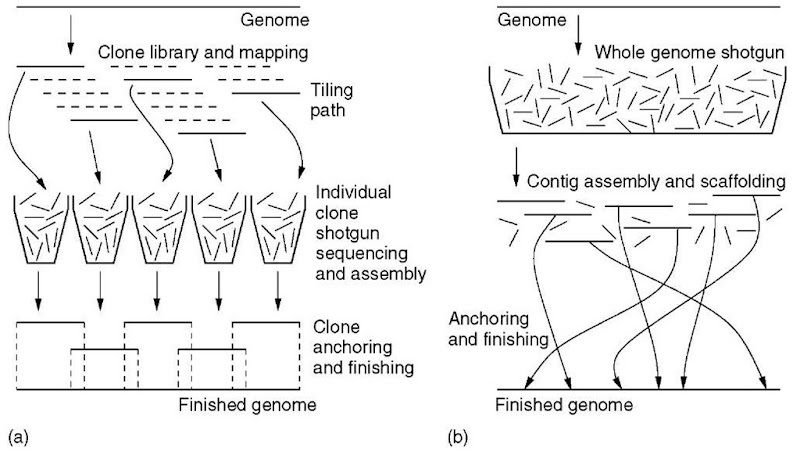
There was considerable debate as to the most desirable technique for sequencing the human genome (Weber and Myers, 1997; Green, 1997), much of which related to the issue of assembly. The publicly funded project favored what has come to be called the clone-by-clone approach (Figure 1a). This method had been used successfully for the preliminary C. elegans project (C. elegans Sequencing Consortium, 1998). Here, a minimal set of large-insert clones, for example, Bacterial Artificial Chromosome (BAC) clones, are sequenced individually and assembled in conjunction with the aid of longer-range data, for example, fingerprint maps and clone end sequences. Investigators were optimistic because the clones and the scale of mapping data are typically larger than the scale of repeats. The latter are then essentially encapsulated, permitting a theoretically unambiguous assembly.
Conversely, a newly founded private venture called Celera favored a whole genome shotgun (WGS) methodology (Myers, 1999). Here, genomic DNA is sheared and sequenced directly (Figure 1b). Although the WGS technique had already been proven on a number of microbial genomes (e.g., Fleischmann et al., 1995), it was unclear whether success in assembling these low-repeat data would extend to the repeat-laden human genome. Proponents based their assessment of feasibility on a number of analyses. In particular, they predicted, based on the Lander and Waterman (1988) theory, that the number of gaps in the sequence would be small (Marshall and Pennisi, 1998). Simulations also suggested that the assembly problem would be tractable, despite the problem of repeats (Weber and Myers, 1997).
Figure 1 Diagrammatic representation of clone-by-clone sequencing (a) and whole genome shotgun sequencing (b)
Ultimately, these two projects would proceed concurrently, but amid some controversy. WGS advocates maintained that the mapping phase of clone-by-clone sequencing was a slow, expensive, and unnecessary step. Conversely, sentiment in the public project was that a pure WGS assembly would not be well positioned for the downstream phase of completing the genome. In the following sections, we describe each of these two methods in greater detail. We will then draw a few comparisons between the results.
3. Whole genome shotgun approach
A purely WGS method does not utilize long-range mapping data or other nonsequence information to generate an assembly. From an algorithmic standpoint, the main design issue becomes correctly detecting and resolving repeated sequence in the native DNA. In their original proposal, Weber and Myers (1997) cited the so-called double-barrel shotgun approach as a mechanism of overcoming repeat difficulties. Here, inserts are size-selected such that their average length is greater than twice the average read length. These inserts can be sequenced from both ends, which yields not only coverage of the target, but also “mating” information for the pair of reads. That is, the separation distance between these reads is known, at least approximately, from the original insert size. As reads start coalescing into contiguous segments, this mate-pair information is useful in linking sequence contigs into larger “scaffolds”. Moreover, it facilitates the encapsulation of repeat structures, as outlined above. This implies the necessity of obtaining mate-pair data over a variety of length scales.
One of the acknowledged liabilities of the WGS method is its susceptibility to errors that arise from corrupted mate-pairing information. Chimerism in clones is always a factor, although this can largely be eliminated by carefully designing and executing protocols. The larger contribution in early WGS projects came from sample tracking errors in the physical handling of DNA and lane tracking errors in analyzing gel images. Overall, such errors meant as many as 10% of the mate pairs were incorrect (Myers, 1999). By the time the pilot WGS project for Drosophila was under way, these mispairings could be reduced to substantially less than 1% (Myers et al., 2000). Consequently, mate-pair information could actively be utilized in the assembly process. On this basis, Myers et al. (2000) developed the assembly software, ultimately used for both the Drosophila and human genomes, which came to be called the Celera assembler.
3.1. Basic assembly methodology
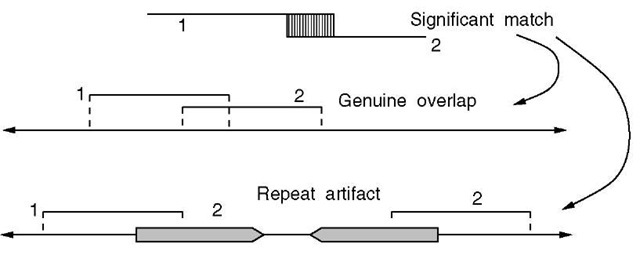
The assembly process nominally begins with a computation that identifies all fragment overlaps. Input is in the form of a set of N fragments that have passed screens for vector and other contamination and that have been appropriately trimmed for base quality. Each new candidate fragment is compared to all existing fragments in the assembly, so that there are a total of CNj2 = N (N — 1)/2 comparisons. Matches of at least 40 bp and 94% similarity were viewed as significant. Such cases represent either a true fragment overlap or a repeat artifact (Figure 2). Misinterpreting the latter case as an overlap would effectively exclude an intervening segment of the original sequence.
In the next phase, sequence “unitigs” are constructed. Unitigs are defined by Myers et al. (2000) as local sequence assemblies whose overlaps are not disputed by any other data. However, this does not preclude the possibility of an erroneously assembled repeat region. Unitigs that exhibit uncharacteristically deep coverage are liable to be overcollapsed repeats. A probabilistic measure called the A-statistic, a, was devised to detect such cases (see Section 3.2). Large a values are associated with a small likelihood of a repeat-induced assembly error. Some of the more tractable repeat structures can be resolved at this point, using a dynamic programming algorithm to deduce repeat boundaries.
All unitigs determined to be free of repeat encumbrance are passed on to the “scaffolding” phase. Here, unitigs associated by mate-pair information are linked into scaffolds. In addition to yielding the order of unitigs, mate-pair data also allow them to be oriented relative to one another. A single mate pair is generally insufficient to declare a link, however, two or more imply a link to a statistically vanishing chance of error (Myers etal., 2000). Longer-range scaffolding is also performed on the basis of BAC end data. At the conclusion of this step, one largely has an ordered, scaffolded assembly of the nonrepetitive portion of the genome.
At this point, a series of increasingly aggressive steps of repeat resolution are undertaken. The first round attempts to place unitigs having relatively small a values and at least two links to other contigs. The next step concentrates on unitigs with only a single link, except that it also requires the placement to be within the context of a tiling of other unitigs. In the original assembler used for the Drosophila project, gaps would be filled with the best overlap tiling of remaining unitigs; however, this step was omitted from the human assembler. At the conclusion of these steps, a consensus sequence is generated.
Figure 2 A significant match between two fragments represents a genuine overlap, or the collapse of a region due to a repeat structure. In the former case, fragments 1 and 2 actually derive from the same location in the source DNA molecule, while in the latter case they do not. If a repeat artifact were to be erroneously accepted as a valid overlap, the intervening region would be lost
3.2. Probabilistic repeat detection
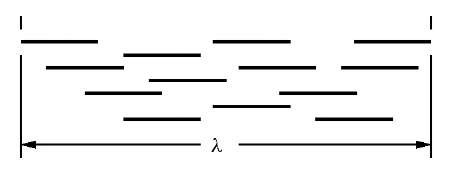
The primary metric used to detect collapsed repeat regions is the so-called A-statistic, denoted here by the symbol a. This statistic is derived according to a Poisson model for the number of reads one expects to find within a contig of length X (Figure 3). Specifically, in a genome of estimated length G, the chance of a single read landing within the boundary of the given unitig is X/G. This approximation neglects end effects, which is justified for large genomes. Presuming the N reads in the project to be independently and identically distributed (IID), one would then expect to find \x = NX/G reads in the average unitig of length X. In other words, in the absence of repeats, there would be an average of about \x reads from all contigs of length X. Because N and X/G are large and small, respectively, the population of reads essentially follows a Poisson process. That is, the probability of finding exactly k reads in a specific unitig of length X is the Poisson probability
where e ^ 2.71828 is Euler’s number.
Now consider a unitig that is actually composed of two regions that have collapsed in the assembly because of a repeat structure (Figure 2). The average such contig would contain 2^ reads, so that the associated probability distribution is P (k, 2^.). The A-statistic is simply the base-10 log of the ratio of these two expressions.
This expression relates the size of a contig to the number of reads it contains. Small values imply that a contig of given size contains more reads than what could reasonably be explained by random chance. One would then infer a collapsed repeat. Extensive simulations indicated that a > 10 almost always indicates the absence of repeat-related assembly error (Myers et al., 2000).
Figure 3 The A-statistic is based on the reads comprising a unitig of length X bases
3.3. The human WGS assemblies
Venter et al. (2001) described two human shotgun assemblies: the whole genome assembly (WGA) and the compartmentalized shotgun assembly (CSA). Both of these assemblies combined native data generated by the WGS procedure, with additional data obtained from the public sequencing project (see Article 24, The Human Genome Project, Volume 3). WGS data comprised about 27 million reads derived from 2-, 10-, and 50-kb inserts. Variation in insert sizes provided linking information over several length scales. Public data were added in the form of 16 million synthesized reads, whose nature remains somewhat controversial (see Section 5). An additional 100 000 BAC end sequences were appropriated for long-range linking information.
The WGA assembly was performed essentially according to the algorithm described above. Parameters were selected such that error likelihood for each sequence overlap was about 10—17. Unitigs were evaluated using equation 2, with the resulting set covering about 74% of the genome. After scaffolding and repeat resolution, BAC data were used to close additional gaps. The resulting assembly consisted of scaffolds spanning about 2.85 Gb with sequence coverage of about 2.59 Gb. There were about 11.3 million “chaff” reads that could not be incorporated into the assembly. Average scaffold and contig sizes were 1.5 Mb and 24 kb, respectively, and the total number of contigs was 221 000.
The CSA assembly is based on the premise of isolating smaller regions of the genome according to BAC contigs from the public project, creating localized assemblies using a combination of WGS and public data, and then tiling these assemblies into a larger result (Huson et al., 2001). A WGA assembly is then created from each resulting component. Here, about 6.2 million reads were ultimately “chaff”. Average scaffold and contig sizes were 1.4Mb and 23.2 kb, respectively, and the total number of contigs was 170 000. Anchoring of scaffolds to chromosomes was undertaken with the aid of physical mapping information from the public sequencing project (McPherson et al., 2001).
4. Clone-by-clone approach
The public sequencing effort explicitly set out with the goal of deciphering the genome in base-perfect form. That is, the error rate should be a maximum of one base in every 10 000. The approach would ideally be one of “divide and conquer” in two sequential steps. On a global level, the genome would be broken into a set of large-insert clones, predominantly BACs (Shizuya et al., 1992). At an average length exceeding 100 kb, they are larger than, and thus isolate most repeat structures. In the original plan, a physical map describing the order of these clones would then be generated via the high-throughput fingerprint technique (Marra et al., 1997). Here, clones are digested to create “fingerprints” of fragment lengths. These lengths are inferred from band positions on electrophoretic gel images. A significant number of fragment length matches between two fingerprints indicate a likely overlap between their associated clones. The map would enable the selection of a minimally overlapping set of clones, each of which would then be locally shotgun sequenced and finished. With the given complement of information, generating the genome-wide consensus sequence would then essentially be a formality.
Investigators in the public effort deemed this approach more suitable than pure WGS on several grounds. First, WGS had not been proven on a repeat-rich genome and there was concern regarding the ability to use a draft assembly as a substrate for obtaining the base-perfect sequence. The outbred nature of the source DNA posed similar concerns. Moreover, previous projects had demonstrated that the mapping phase was a comparatively inexpensive component, usually less than 10% of total cost (Green, 1997). Improved mapping techniques had also been developed to increase throughput (Marra et al., 1997). The intermediate resolution afforded by BAC clones would also allow better targeting of underrepresented regions caused by inevitable cloning biases. Lastly, it would be more compatible with the distributed and international nature of the project.
A pilot phase proved the general feasibility of this approach by finishing a small portion of the genome (Lander et al., 2001). Around the same time, an alternative proposal was made to first generate a genome-wide draft sequence, while deferring the finishing phase to a later date. This was driven by two considerations. First, the research community was eager to obtain sequence data as quickly as possible. Second, the commercially funded Celera project sparked strong concerns about the privatization of the human genome and the rise of proprietary human databases (Pennisi, 1999). The public project thus proceeded along a modified version of the clone-by-clone approach as described below.
4.1. Clone selection
The modified project schedule actually necessitated constructing the physical map in parallel with the clone-sequencing phase rather than beforehand (McPherson et al., 2001). Consequently, clones could not be solely selected with guidance from a completed map to achieve a minimum tiling path. They were initially selected on the basis of having no overlap with already-sequenced clones or those in the sequencing pipeline. These “seed” clones formed a nonoverlapping set and thus functioned as nucleation sites for sequencing distinct regions of the genome. The fidelity of candidate seed clones was checked by comparing fingerprint bands to neighboring clones, to the clone size, and to its corresponding map fingerprint. Clones from contig ends were explicitly excluded because of their limited ability to confirm bands.
A clone registry was created for managing clones claimed by various participants for sequencing. Seed clones were then extended by picking mating clones having relatively high overlap scores (Sulston et al., 1988). Band confirmation and BAC end data were also used in this capacity. In cases where a seed clone sequence was available, overlaps could be further confirmed by comparison to the end sequence of the candidate extension clone. The average overlap in the final clone set was found to be 47.5 kb (about 28%).
4.2. Processing individual clones
Individual large-insert clones were sequenced according to the standard shotgun methodology. Clones were typically sonicated and the resulting fragments were size-selected and subcloned into M13 or plasmid vectors. Details varied among the participants, for example, the proportions in which dye-primer and dye-terminator reactions were used and the type of sequencing platforms that were employed. Shotgun coverage was typically obtained to at least sixfold depth. Reads were assembled predominantly by the Phrap software package (Gordon etal., 1998), which uses an overlap-layout-consensus algorithm.
As with the WGS procedure, repeats tend to cause collapsed regions of misas-sembled reads. However, the localized nature of the assembly decreases the repeat problem substantially. Sequencing errors further complicate the process, which Phrap addresses through base-quality values assigned at the upstream base-calling step (Ewing et al., 1998). Phrap does not exploit mate-pair information in the way the Celera assembler does. Moreover, reads are predominantly derived from the same clone source, reducing problems associated with sequence polymorphism.
The first step of the Phrap assembly process determines which reads overlap by evaluating all pairwise subclone comparisons. Anomalies, such as unremoved vector and chimeric reads, are identified as well. Revised quality scores are then computed for each base. These reflect both the raw base-quality scores from the individual reads and any enhancements based on mutual overlap confirmation. The second step determines the optimal placement of overlapping reads to form a layout of the assembly. Here, sequence alignment scores are used in decreasing order to merge contigs. Some joints are rearranged at this stage as well. Finally, consensus sequence is generated for each base position from the columnized layout.
4.3. Creating a draft assembly
Assembly at the genome level involves combining the information from the clone overlap physical maps and validating those overlaps using the sequence. Given the draft nature of most of the clones and the fact that some were not yet represented in the map, this phase would not be trivial. After a uniform filtering procedure for contaminated sequences, the assembly approach proceeded in two steps: layout and merging.
The layout step sought to associate sequenced clones with clones on the physical map. Ideally, this would simply be a matter of mapping the names between the two clone sets. However, to minimize mistakes at this stage, two additional quality control procedures were instituted. Fingerprint accuracy was such that in silico digests could be used to confirm associations for sequence clones that had few enough contigs. BAC end sequences were also used in a similar capacity. A total of 25 403 clones were linked in this fashion. Fingerprint clone contigs were localized to their respective chromosomal locations using fluorescence in situ hybridization (FISH) data (see Article 22, FISH, Volume 1) and a variety of radiation hybrid and genetic maps (Lander et al., 2001).
Software was then used in the merge phase to order and orient the entire collection of sequenced clones (Kent and Haussler, 2001). Sequence clones were initially aligned to those within the same fingerprint clone contig. Groups of overlapping clones were then built up into larger ordered “barges”. Order and orientation were further pursued with the aid of additional information, for example, mRNAs, STSs, BAC end reads, and any other available linking information. A sequence path was then generated from the resulting structure.
The two-phase process used for the public sequencing effort resulted in the draft assembly described by Lander et al. (2001). They reported an overall N50 length for sequenced clone contigs of 826 kb. This is the length of the contig in which the average nucleotide resides. The N50 statistic varied by chromosome. In particular, it was a few orders of magnitude higher for chromosomes 21 and 22, which had already been completed (Dunham etal., 1999; Hattori etal., 2000). The overall assembly consisted of 4884 sequenced clone contigs and suggested a euchromatic genome size of 2.9 Gb.
5. Epilogue
The public and private genome assemblies were published simultaneously in February of 2001 (Lander et al., 2001; Venter et al., 2001). Celera’s results quickly came under scrutiny based partially upon the amount of public sequencing data that had been appropriated, but more so on the manner in which it had been used. Waterston etal. (2002a) described the apparently nonrandom fashion in which synthetic “reads” were manufactured from the public genome assembly and tiled over Celera’s own data. They further clarified the extent to which public marker and map data were used by Celera and the nature of the CSA assembly, which was the basis of all the biological analysis reported by Venter et al. (2001). Emphasizing the fact that Celera did not report an assembly comprised solely of their own shotgun data, Waterston etal. concluded that the published Celera assemblies did not represent a legitimate proof of concept of the WGS method applied to a complex genome. This view was echoed by other members of the scientific community (Green, 2002). Myers et al. (2002) rebutted this position, asserting that no linking information was retained in the synthetic reads and that the WGA and CSA assemblies were, in fact, pure WGS assemblies. Vigorous debate continues (Waterston et al., 2003; Adams et al., 2003; Istrail et al., 2004).
It will likely be some time yet before history conclusively places these efforts into a more objective relational context. Celera would certainly have averted any claims by simply eschewing public data. Their initial projections regarding the difficulty of such an assembly based on shotgun data alone, for example, number of gaps and amount of chaff, were clearly incorrect. This created an unexpected urgency for more data. While their use of public data was unquestionably legitimate, it has significantly blurred the comparison between these two fundamentally different approaches.
Regardless, the Celera assemblies might ultimately be relegated to scientific footnotes in the sense that they are unlikely to be extended into finished human sequences. The drive toward completing the genome is proceeding solely on the basis of the public data, and chromosomes are now being individually finished in fairly rapid succession (Deloukas etal., 2001; Heilig etal., 2003; Hillier etal., 2003; Mungall etal., 2003; Dunham etal., 2004; Grimwood etal., 2004). It is somewhat ironic that neither the WGS nor the clone-by-clone strategies appear likely to carry forward in pure form for future projects. Investigators currently favor hybrid approaches that combine aspects of both these techniques (Waterston et al., 2002b). The art of sequencing and assembling genomes is clearly an evolving one.