1. Definition
Some protein domains are composed of units of similar structure (see Figure 1). Often, but not always, these units are also similar in sequence, which explains why they have a similar structure. Therefore, they can be considered protein repeats that originated by duplications from a single ancestral sequence.
These small units are large enough to form secondary structural elements but too small to be stable by themselves. They acquire stability by folding together in a repetitive structure. This is the defining feature of protein repeats. They occur repeated, never alone.
Repeats tend to be tandemly repeated (one next to the other in sequence), but the occurrence of insertions of variable length in between repeats is sometimes observed. Those insertions tend to lack structure and to be short, although there seems to be no obvious reason for that. We can speculate that because folding of a structure composed of repeats is more delicate than that of globular folds, large insertions would make the fold unstable or difficult to form.
Since repeats are consecutive in sequence, a condition on their structure is that the C-terminal must be spatially near the N-terminal in order to produce units that can fold together in a compact continuum of repeat units. Given the fact that the elements of secondary structure are quite linear, a repeat must contain a minimum of two of them to bring the C-terminal of the repeat close to the N-terminal. This imposes a minimum length of about 12 amino acids on these repeats. An upper bound of about 50 amino acids can be explained because a larger unit could fold into an independent domain that does not need another repeat to form a stable fold.
2. Classification
Repeats can be classified, like protein folds, depending on the secondary structural elements that compose them. Accordingly, most widespread repeats can be grouped in three groups, all-alpha, all-beta, and alpha/beta:
• Armadillo/HEAT repeats and tetratricopeptide repeats (TPR-like) in the all-alpha group.
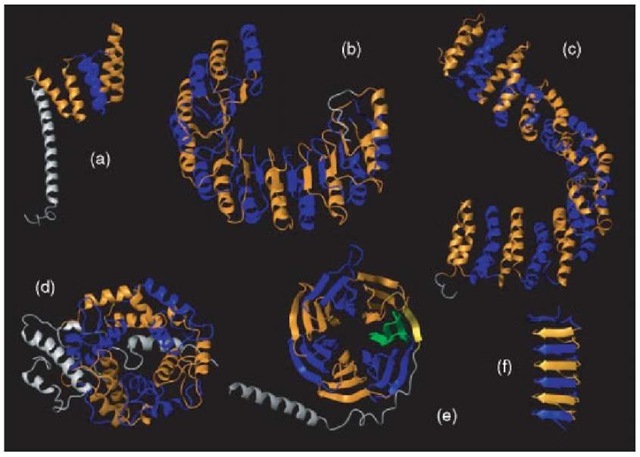
Figure 1 Examples of protein domains composed of repeats. (a) TPR repeats (PDB id: 1a17). (b) Leucine-rich repeats (PDB id: 1dfj, chain I). (c) HEAT repeats (PDB id: 1b3u). (d) PFTA repeats (PDB id: 1ft2, chain A). (e) WD40 repeats (PDB id: 1gp2, chain B). (f) Beta-loop repeats in an antifreeze protein (PDB id: 1ezg). Blue and orange represent alternating repeats. The rest of the structure is colored in grey. In the case of WD40 (e), one of the repeats is composed of a beta strand (in yellow) previous to the first repeat and three C-terminal beta strands (in green); this way of closing a barrel composed of beta-sheets is observed in other protein families of similar structure. The figure was composed using the MOLMOL software (Koradi et al., 1996)
• Beta propellers (as WD40 and Kelch) and beta trefoils in the all-beta group.
• Leucine-rich repeats (LRR) and ankyrin repeats in the alpha/beta group.
There are many more repeat families that occur less frequently (Andrade et al., 2001a), such as the beta-loop 12-residue repeats found in an insect antifreeze protein.
The shape of the superstructure can also define the function and dynamics of the repeat unit and, therefore, must also be taken into account. Repeats can form open structures (see examples in Figures 1a, 1b, 1c, and 1f) or closed structures (see examples in Figures 1d and 1e). Open structures formed by repeats are observed with variable copy number, whereas close structures are more constrained to a copy number appropriate to the closing of the superstructure (Andrade et al., 2000).
3. Structure and function
In general, domains composed of repeats confer to proteins an enlarged binding surface area with the possibility of multiple binding and structural roles. This is the reason protein-protein interaction is the most prevalent function of proteins with repeats (Andrade et al., 2001b). The periodicity of the three-dimensional structure favors spatially periodic or symmetric interactions that are critical for structural integrity in various cellular contexts (for example, spectrin in the submembrane lamina or HEAT repeats in the vesicular coat structures). Among other functions performed by proteins with repeats, we can find enzymatic activities, ice binding (in antifreeze proteins), or energy storage proteins in plant seeds.
4. Evolution and distribution
Repeats have appeared multiple times along evolution. They originate by duplication and recombination within a single gene. The evolution of repeats from a common ancestor that necessarily must have contained a single repeat seems paradoxical, given that apparently more than one repeat is required for folding. One possible explanation is that the ancestor formed homo-oligomers where two identical chains provided the repetition (Ponting et al., 2000).
It is estimated that 14% of all proteins have repeats (Marcotte et al., 1999). Their actual number is probably higher, because the estimate is based on sequence comparison, and therefore missing those that have no detectable sequence similarity.
Repeats are present over all taxa but are more abundant in eukaryotic organisms and among them most frequent in metazoans. This distribution shows a correlation between the increase of organism complexity and the already mentioned principal function of domains composed of repeats, binding proteins.
5. Detection
Identifying tandem repeats with high sequence similarity is relatively easy. However, it happens that the constraints in sequence conservation among repeats are relatively lax. For example, the HEAT repeats of the structure displayed in Figure 1c average only 18% of sequence identity. Their short length complicates their detection too, and the number of repeats per protein may differ between members of the same family. In the absence of a reference structure, it is especially difficult to detect the boundaries of each repeated unit. Several methods have been published dealing with the detection and analysis of repeats (REP, http://www.embl.de/~andrade/papers/rep/search.html, Andrade et al., 2000; REPRO, http: //ibivu.cs.vu.nl/programs/reprowww/, Heringa, 2000; Pellegrini etal., 1999).