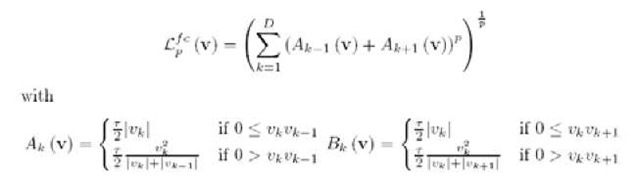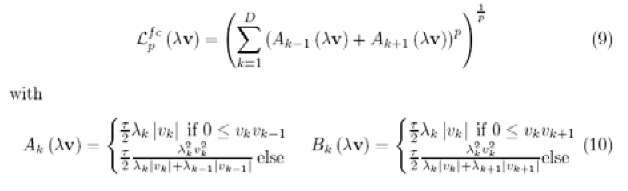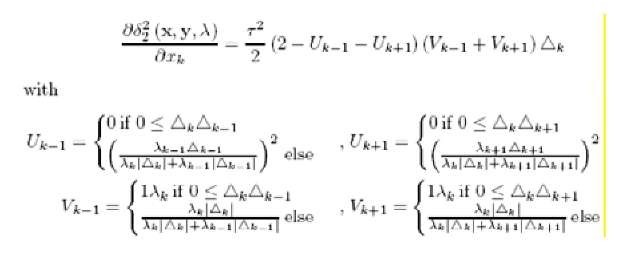INTRODUCTION
Bioinformatics has become an important tool to support clinical and biological research and the analysis of functional data, is a common task in bioinformatics (Schleif, 2006). Gene analysis in form of micro array analysis (Schena, 1995) and protein analysis (Twyman, 2004) are the most important fields leading to multiple sub omics-disciplines like pharmacogenomics, glyco-proteomics or metabolomics. Measurements of such studies are high dimensional functional data with few samples for specific problems (Pusch, 2005). This leads to new challenges in the data analysis. Spectra of mass spectrometric measurements are such functional data requiring an appropriate analysis (Schleif, 2006). Here we focus on the determination of classification models for such data. In general, the spectra are transformed into a vector space followed by training a classifier (Haykin, 1999). Hereby the functional nature of the data is typically lost. We present a method which takes this specific data aspects into account. A wavelet encoding (Mallat, 1999) is applied onto the spectral data leading to a compact functional representation. Subsequently the Supervised Neural Gas classifier (Hammer, 2005) is applied, capable to handle functional metrics as introduced by Lee & Verleysen (Lee, 2005). This allows the classifier to utilize the functional nature of the data in the modelling process. The presented method is applied to clinical proteome data showing good results and can be used as a bioinformatics method for biomarker discovery.
BACKGROUND
Applications of mass spectrometry (ms) in clinical proteomics have gained tremendous visibility in the scientific and clinical community (Villanueva, 2004) (Ketterlinus, 2005). One major objective is the search for potential classification models for cancer studies, with strong requirements for validated signal patterns (Ransohoff, 2005). Primal optimistic results as given in (Petricoin, 2002) are now considered more carefully, because the complexity ofthe task of biomarker discovery and an appropriate data processing has been observed to be more challenging than expected (Ransohoff, 2005). Consequently the main recent work in this field is focusing on optimization and standardisation. This includes the biochemical part (e.g. Baumann, 2005), the measurement (Orchard, 2003) and the subsequently data analysis (Morris, 2005)(Schleif 2006).
PROTOTYPE BASED ANALYSIS IN CLINICAL PROTEOMICS
Here we focus on classification models. A powerful tool to achieve such models with high generalization abilities is available with the prototype based Supervised Neural Gas algorithm (SNG) (Villmann, 2002). Like all nearest prototype classifier algorithms, SNG heavily relies on the data metric d, usually the standard Euclidean metric. For high-dimensional data as they occur in proteomic patterns, this choice is not adequate due to two reasons: first, the functional nature of the data should be kept as far as possible. Second the noise present in the data set accumulates and likely disrupts the classification when taking a standard Euclidean approach. A functional representation of the data with respect to the used metric and a weighting or pruning of especially (priory not known) irrelevant function parts of the inputs, would be desirable. We focus on a functional distance measure as recently proposed in (Lee, 2005) referred as functional metric. Additionally a feature selection is applied based on a statistical pre-analysis ofthe data. Hereby a discriminative data representation is necessary. The extraction of such discriminant features is crucial for spectral data and typically done by a parametric peak picking procedure (Schleif, 2006). This peak picking is often spot of criticism, because peaks may be insufficiently detected and the functional nature of the data is partially lost. To avoid these difficulties we focus on a wavelet encoding. The obtained wavelet coefficients are sufficient to reconstruct the signal, still containing all relevant information of the spectra, but are typically more complex and hence a robust data analysis approach is needed. The paper is structured as follows: first the bioinformatics methods are presented. Subsequently the clinical data are described and the introduced methods are applied in the analysis of the proteome spectra. The introduced method aims on a replacement of the classical three step procedure of denoising, peak picking and feature extraction by means of a compact wavelet encoding which gives a more natural representation of the signal.
BIOINFORMATIC METHODS
The classification ofmass spectra involves in general the two steps peak picking to locate and quantify positions of peaks and feature extraction from the obtained peak list. In the first step a number of procedures as baseline correction, denoising, noise estimation and normalization are applied in advance. Upon these prepared spectra the peaks have to be identified by scanning all local maxima. The procedure of baseline correction and recalibration (alignment) of multiple spectra is standard, and has been done here using ClinProTools (Ketterlinus, 2006). As an alternative we propose a feature extraction procedure preserving all (potentially small) peaks containing relevant information by use of the discrete wavelet transformation (DWT).Due to the local analysis property of wavelet analysis the features can still be related back to original mass position in the spectral data which is essential for further biomarker analysis. For feature selection the Kolmogorov-Smirnoff test (KS-test) (Sachs, 2003) has been applied. The test was used to identify features which show a significant (p < 0.01) discrimination between the two groups (cancer, control). In (Waagen, 2003) also a generalization to a multiclass experiment is given. The now reduced data set has been further processed by SNG to obtain a classification model with a small ranked set of features. The whole procedure has been cross-validated in a 10-fold cross validation.
WAVELET TRANSFORMATION IN MASS SPECTROMETRY
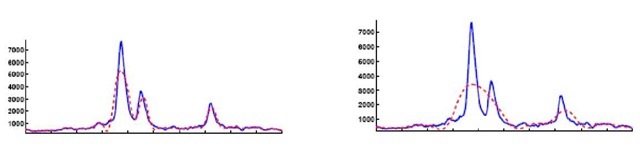
Wavelets have been developed as powerful tools (Rie-der, 1998) used for noise removal and data compression. The discrete version of the continuous wavelet transform leads to the concept of a multi-resolution analysis (MRA). This allows a fast and stable wavelet analysis and synthesis. The analysis becomes more precise if the wavelet shape is adapted to the signal to be analyzed. For this reason one can apply the so called bi-orthogonal wavelet transform (Cohen, 1992), which uses two pairs of scaling and wavelet functions. One is for the decomposition/analysis and the other one for reconstruction/synthesis, giving a higher degree of freedom for the shape of the scaling and wavelet function. In our analysis such a smooth synthesis pair was chosen. It can be expected that a signal in the time domain can be represented by a small number of a relatively large set of coefficients from the wavelet domain. The spectra are reconstructed in dependence of a certain approximation level L of the MRA. The denoised spectrum looks similar to the reconstruction as depicted in Figure 1.
One obtains approximation- and detail-coefficients (Cohen, 1992). The approximation coefficients describe a generalized peak list, encoding primal spectral information. For linear MALDI-TOF spectra a device resolution of500-800L>a can be expected. This implies limits to the minimal peak width in the spectrum and hence, the reconstruction level of the Wavelet-Analysis should be able to model corresponding peaks. A level L = 4 is appropriate for our problem (see Figure 1). Applying this procedure including the KS-test on the spectra with an initial number of 22306 measurement points per spectrum one obtains 602 wavelet coefficients used as representative features per spectrum, still allowing a reliable functional representation of the data. The coefficients were used to reconstruct the spectra and the final functional representation of the signal.
Figure 1. Wavelet reconstruction of the spectra with L = 4, 5, x-mass positions, y-arbitrary unit. Original signal – solid line. One observes for L = 5 (right plot) the peak approximate is to rough.
PROTOTYPE CLASSIFIERS
Supervised Neural Gas (SNG) is considered as a representative for prototype based classification approaches as introduced by Kohonen (Kohonen, 1995). Different prototype classifiers have been proposed so far (Kohonen, 1995) (Sato, 1996) (Hammer, 2005) (Villmann, 2002) as improvements of the original approach. The SNG has been introduced in (Villmann, 2002) and combines ideas from the Neural Gas algorithm (NG) introduced in (Martinetz, 1993) with the Generalized learning vector quantizer (GLVQ) as given in (Sato, 1996).
Subsequently we give some basic notations and remarks to the integration of alternative metrics into Supervised Neural Gas (SNG). Details on SNG including convergence proofs can be found in (Villmann, 2002). Let us first clarify some notations: Let cv in L be the label of input v, L a set of labels (classes). Let V in RDV be a finite set of inputs v. LVQ uses a fixed number of prototypes for each class. Let W={wr} be the set of all codebook vectors and c be the class label of w . Furthermore, let Wc=/wr/c = c} be the subset of prototypes assigned to class c in L. The task of vector quantization is realized by the map W as a winner-take-all rule, i.e. a stimulus vector v in Vis mapped onto that prototype s the pointer ws of which is closest to the presented stimulus vector v, measured by a distance dx (v,w). dx (v,w) is an arbitrary differentiable similarity measure which may depend on a parameter vector X. For the moment we take X as fixed. The neuron s (v) is called winner or best matching unit. If the class information of the weight vector is used, the above scheme generates decision boundaries for classes (details in (Villmann, 2002)). A training algorithm should adapt the prototypes such that for each class c in L, the corresponding codebook vectors Wc represent the class as accurately as possible. Detailed equations and cost function for SNG are given in (Villmann, 2002). Here it is sufficient to keep in mind that in the cost function of SNG the distance measure can be replaced by an arbitrary (differentiable) similarity measure, which finally leads to new update formulas for the gradient descent based prototype updates.
Incorporation of a functional metric to SNG As pointed out before, the similarity measure dx (v,w) is only required to be differentiable with respect to X and w. The triangle inequality has not to be fulfilled necessarily (Hammer, 2005). This leads to a great freedom in the choice of suitable measures and allows the usage of non-standard metrics in a natural way. For spectral data, a functional metric would be more appropriate as given in (Lee, 2005). The obtained derivations can be plugged into the SNG equations leading to SNG with a functional metric, whereby the data are functions represented by vectors and, hence, the vector dimensions are spatially correlated. Common vector processing does not take this spatial order of the coordinates into account. As a consequence, the functional aspect of spectral data is lost. For proteome spectra the order of signal features (peaks) is due to the nature of the underlying biological samples and the measurement procedure. The masses of measured chemical compounds are given ascending and peaks encoding chemical structures with a higher mass follow chemical structures with lower masses.
In addition, multiple peaks with different masses may encode parts of the same chemical structure and, hence, are correlated. Lee proposed an appropriate norm with a constant sampling period t:
are respectively of the triangles on the left and right sides of x.. Just as for Lp, the value of p is assumed to be a positive integer. At the left and right ends of the sequence, xg and xD are assumed to be equal to zero. The derivatives for the functional metric taking p = 2 are given in (Lee, 2005). Now we consider the scaled functional norm where each dimension (0, 1], v. is scaled by a parameter X. > 0 and all X. sum up to 1:
The prototype update changes to:
And ifc = J’k – yt using this parameterization one can emphasize/neglect different parts of the function for classification.
ANALYSIS OF PROTEOMIC DATA
The proposed data processing scheme is applied to clinical ms spectra taken from a cancer study (45 cancer, 50 control samples). Sample preparation and profile spectra analysis were carried out using the CLINPROT system (Bruker Daltonik, Bremen, Germany [BDAL]). The preprocessed set of spectra and the corresponding wavelet coefficients are then analyzed using the SNG extended by a functional metric. We reconstructed the spectra based upon the discriminative wavelet coefficients determined by the Kolmogorov-Smirnoff test as explained above and used corresponding intensities as features. We used all features for the parameterized functional norm i.e. all X. = 1. The original signal with approx. 22000 sampling points had been processed with only 600 remaining points still encoding the significant parts of the signal relevant for discrimination between the classes. The SNG classifier with functional metric obtains a crossvalidation accuracy of 84% using functional metric and 82% by use of standard Euclidean metric. The results from the wavelet processed spectra are slightly better than using standard peak lists, with 81% crossvalidation accuracy.
FUTURE TRENDS
The proposed method generates a compact but still complex functional representation of the spectral data. While the bior3.7 wavelet gives promising results they are still not optimal, due to signal oscillations, leading to negative intensities in the reconstruction. Further, the functional nature of the data motivates the usage of a functional data representation and similarity calculation but there are also spectra regions encoded which do not contain meaningful biological information but measurement artefacts. In principle it should be possible to remove this overlaying artificial function from the real signal. Further it could be intere sting to incorporate additional knowledge about the peak width, which is increasing over the mass axis.
CONCLUSION
The presented interpretation of proteome data demonstrate that the functional analysis and model generation using SNG with functional metric in combination with a wavelet based data pre-processing provides an easy and efficient detection of classification models. The usage of wavelet encoded spectra features is especially helpful in detection of small differences which maybe easily ignored by standard approaches as well as to generate a significant reduced number of points needed in further processing steps. The signal must not be shrinked to peak lists but could be preserved in its functional representation. SNG was able to process high-dimensional functional data and shows good regu-larization. By use ofthe Kolmogorov-Smirnofftest we found a ranking ofthe features related to mass positions in the original spectrum which allows for identification of most relevant feature dimensions and to prune irrelevant regions of the spectrum. Alternatively one could optimize the scaling parameters of the functional norm directly during classification learning by so called relevance learning as shown in (Hammer, 2005) for scaled Euclidean metric. Conclusively, wavelet spectra encoding combined with SNG and a functional metric is an interesting alternative to standard approaches. It combines efficient model generation with automated data pre-treatment and intuitive analysis.
KEY TERMS
Bioinformatics: Generic term of a research field as well as a set of methods used in computational biology or medicine to analyse multiple kinds of biological or clinical data. It combines the disciplines of computer science, artificial intelligence, applied mathematics, statistics, biology, chemistry and engineering in the field of biology and medicine. Typical research subj ects are problem adequate data pre-processing of measured biological sample information (e.g. data cleaning, alignments, feature extraction), supervised and unsupervised data analysis (e.g. classification models, visualization, clustering, biomarker discovery) and multiple kinds of modelling (e.g. protein structure prediction, analysis of expression of gene, proteins, gene/protein regulation networks/interactions) for one or multidimensional data including time series. Thereby the most common problem is the high dimensionality of the data and the small number of samples which in general make standard approach (e.g. classical statistic) inapplicable.
Biomarker: Mainly in clinical research one goal of experiments is to determine patterns which are predictive for the presents or prognosis of a disease state, frequently called biomarker. Biomarkers can be single or complex (pattern) indicator variables taken from multiple measurements of a sample. The ideal biomarker has a high sensitivity, specificity and is reproducible (under standardized conditions) with respect to control experiments in other labs. Further it can be expected that the marker is vanishing or changing during a treatment of the disease.
Clinical Proteomics: Proteomics is the field of research related to the analysis of the proteome of an organism. Thereby, clinical proteomics is focused on research mainly related to disease prediction and prognosis in the clinical domain by means of proteome analysis. Standard methods for proteome analysis are available by Mass spectrometry.
Mass Spectrometry: An analytical technique used to measure the mass-to-charge ratio of ions. In clinical proteomics mass spectrometry can be applied to extract fingerprints of samples (like blood, urine, bacterial extracts) whereby semi-quantitative intensity differences between sample cohorts may indicate biomarker candidates
Prototype Classifiers: Are a specific kind ofneural networks and related to the kNN classifier. The classification model consists of so called prototypes which are representatives for a larger set of data points. The classification is done by a nearest neighbour classification using the prototypes. Nowadays prototype classifiers can be found in multiple fields (robotics, character recognition, signal processing or medical diagnosis) trained to find (non)linear relationships in data.
Relevance Learning: A method, typically used in supervised classification, to determine problem specific metric parameter. With respect to the used metric and learning schema univariate, correlative and multivari-ate relations between data dimensions can be analyzed. Relevance learning typically leads to significantly improved, problem adapted metric parameters and classification models.
Wavelet Analysis: Method used in signal processing to analyse a signal by means of frequency and local information. Thereby the signal is encoded in a representation of wavelets, which are specific kinds of mathematical functions. The Wavelet encoding allows the representation of the signal at different resolutions, the coefficients contain frequency information but can also be localized in the signal.