Further Evidence that [URE3] Is a Prion of Ure2p
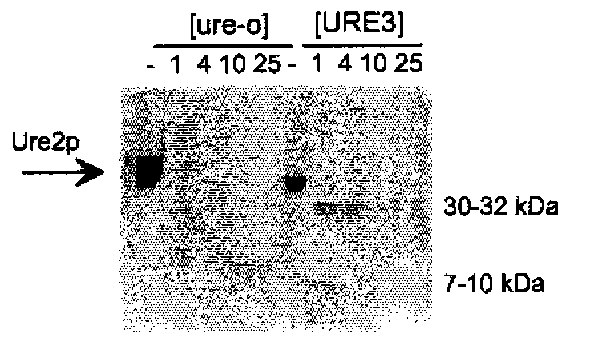
The prion model for [URE3] predicts that the Ure2 protein should be altered in some way in [URE3] strains, compared to wild-type strains. It was shown that, indeed, Ure2p is more protease-resistant in extracts of [URE3] cells than in extracts of (ure-o) cells (Fig. 5; 47). This protease resistance is not simply a general concomitant of derepressed nitrogen regulation, but is a special feature of [URE3] strains (48).
It was important to show that [URE3] is truly arising de novo, and is not a mutant form of a nucleic acid replicon already present in the cell. Such a repli-con would depend on the same chromosomal genes as does [URE3], but would have inactivated functional genes. Thus, a ure2 mutant would have lost this replicon, and would no longer be able to give rise to [URE3] derivatives, even if the URE2 gene were subsequently replaced. In fact, starting with a ure2A strain, replacing the URE2 gene on a plasmid made the strain capable of giving rise to [URE3] derivatives, ruling out this model (48). Spontaneous generation of prions does happen.
It was stated above that overproduction of Ure2p induces the de novo appearance of [URE3], and it was critical to show that, in these experiments, it was the protein, not the overproduced URE2 mRNA or the URE2 gene itself in high copy number, that was making [URE3] appear. When URE2 is placed under the control of the inducible GAL1 promoter, formation of [URE3] is only induced when the promoter is turned on, so that the high copy number of the gene is not the critical factor (7). Removing a single nucleotide from codon 40 of URE2, and introducing a compensatory nucleotide in codon 80, alters the reading frame of most of the prion domain, and produces a protein of normal size with full nitrogen regulation activity, but no prion-inducing activity whatsoever, and normal levels of URE2 mRNA (Fig. 6; 48). Replacing the single nucleotide removed in codon 40 raises prion-inducing activity to a high level, without changing the mRNA levels. That it is the reading frame alteration that is critical here, and not this particular base in codon 40, is shown by lack of effect of removing the entire codon 40 from the full-length gene or from the prion domain alone (48,49). This proves that it is the Ure2 protein whose overproduction results in the de novo formation of the [URE3] inheritable element.
Fig. 5. Protease resistance of Ure2p in (URE3) prion-containing cells. Ure2p is more proteinase K resistant in extracts of (URE3) cells than in extracts of wild-type cells, indicating that Ure2p is altered in (URE3) strains (47). Furthermore, the pattern of protease-resistant fragments is similar to that produced from the amyloid form of Ure2p produced in vitro (66). The figure shows a Western blot of extracts probed with antibody to the prion domain of Ure2p.
Positive regulatory circuits can produce a nonchromosomal genetic element, and Ure2p is a regulator of transcription (via its effects on Gln3p). However, it is clear that this is not the mechanism of [URE3]. The prion domains and nitrogen regulation domains of Ure2p are separable (Fig. 4; 47) , and the propagation of [URE3] does not depend on the repressed or derepressed state of nitrogen catabolism (48).
Further Evidence that [PSI] Is a Prion of Sup35p
A nonsense mutation in the prion domain of SUP35 has little effect on mRNA levels, but eliminates the prion-inducing activity of the overexpressed gene, indicating that it is the protein, not the mRNA, that is inducing [PSI]s appearance (51).
One nonprion model for [PSI] was a self-propagating suppression, in which expression of a suppression-promoting protein itself required suppression. This model is ruled out by the lack of correlation of suppression level with propagation of [PSI] (46). Expression of the Sup35p prion domain as a separate molecule from the C-terminal domain allows propagation of [PSI] in an antisuppressing environment.
The Sup35 protein was found to be aggregated specifically in extracts of [PSI+] strains (60), an aggregation reflecting in vivo aggregation of the protein, as shown by using a Sup35-GFP fusion protein (61).
Fig. 6. The Ure2 protein induces de novo [URE3] formation. Overproduction of Ure2p from an inducible GAL1 promoter results in an increased frequency with which [URE3] arises (7). Adding one nucleotide in codon 80 of URE2 produces a prion domain fragment with greatly elevated [URE3]-inducing activity (48). Further removing one nucle-otide in codon 44 changes the reading frame of most of the prion domain, eliminating [URE3]-inducing activity, without changing the level of mRNA (48,54). It was confirmed that the nucleotide removed from codon 44 was not of special importance, by removing the entire codon 44; this did not affect prion-inducing activity (49) .
Self-Promoting Filament Formation by Sup35p
The genetic properties of [PSI] and SUP35 indicate that [PSI] is a prion form of Sup35p, but do not indicate what is the mechanism of the prion. Several biochemical studies suggest that [PSI] is an amyloid form of Sup35p whose self-catalysis of amyloid formation is the basis of propagation of the [PSI] prion.
The aggregates of Sup35p, found specifically in extracts of (PSI+) strains, were found to be self-propagating (62). Addition of [PSI+] extracts, containing aggregated Sup35p, to extracts of [psi-] cells, whose Sup35p was mostly soluble, resulted in the conversion of the soluble Sup35p to the aggregated form (62) .
In a different approach, King et al. (63) showed that Sup35p2114 made in Escherichia coli, spontaneously formed filaments in 40% acetonitrile, at pH 2.0. These filaments were high in p-sheet structure, had slight protease resistance, and showed clear birefringence on staining with Congo red, a staining property characteristic of amyloid.
Glover et al. (64) made full-length Sup35p, as well as several fragments, in E. coli, and found that all fragments containing the prion domain spontaneously formed filaments in vitro. The full-length Sup35p formed filaments after a long delay, which could be shortened by addition of either preformed filaments or by addition of an extract of a [PSI+] strain. All of the filaments formed were high in p-sheet content, and those formed from the prion domain and middle domain showed birefringence on staining with Congo red (S. Lindquist, personal communication).
Amyloid Formation by Ure2p Promoted by the Prion Domain of Ure2p
The genetic studies described above indicate that [URE3] is a prion of Ure2p, a self-propagating inactive altered form of the Ure2 protein. The first hint of the mechanism by which [URE3] is self-propagating was the finding that Ure2p is relatively proteinase K-resistant in extracts of strains carrying [URE3], compared to wild-type strains (47). The Ure2p in extracts of wild-type strains was rapidly digested, but Ure2p in extracts of [URE3] cells gave rise to transient 30 and 32 kDa species, and to a longer-lived 7-10 kDa species (Fig. 5). The specificity of the antibody used in the Western blots done for this experiment indicated that the 7-10 kDa species is mostly derived from the N-terminal prion domain of Ure2p (47).
Using Ure2-GFP fusion genes, it was found that Ure2p is aggregated specifically in [URE3] cells (Fig. 7; 65). This in vivo aggregation required the prion domain of Ure2p, as well as the presence of the [URE3] prion. In [ure-o] cells, the Ure2-GFP fusion protein was evenly distributed in the cytoplasm (65).
Fig. 7. Aggregation of Ure2p in [URE3] cells. A Ure2p-green fluorescent protein fusion protein, which was both active in nitrogen regulation and capable of involvement in the prion change, was expressed in wild-type, [URE3] prion-containing, and cured cells. In normal cells, the fusion protein was evenly distributed throughout the cytoplasm, but in prion-containing cells it was aggregated (65).
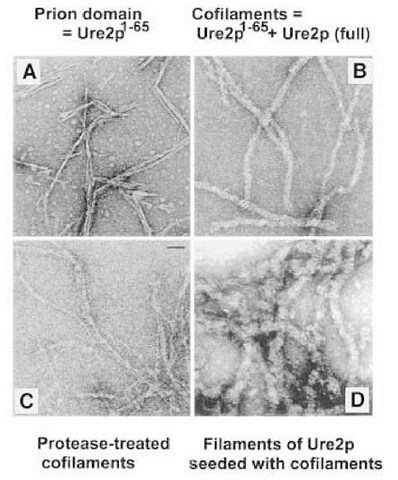
Taylor et al. (66) found that synthetic Ure2p1-65 spontaneously formed amyloid filaments in vitro (Fig. 8). These 45A filaments had all of the characteristics of classic amyloid: They were filaments that were high in p-sheet content, protease-resistant, and showed the birefringence, on staining with Congo red, characteristic of amyloid (Fig. 9; 66). Moreover, these filaments formed at physiological pH and salt conditions.
Under conditions in which the full-length native Ure2p purified from yeast is stably soluble, addition of equimolar amounts of the Ure2p1-65 peptide leads to the formation of cofilaments 200A in diameter, consisting of equimolar amounts of the Ure2p1-65 peptide and full-length Ure2p. This cofilament formation is highly specific, in that Ure2p1-65 does not form cofilaments with proteins other than Ure2p. Moreover, Ap1-42, which also forms amyloid in vitro, does not form cofilaments with Ure2p (66). These cofilaments also have all the properties of amyloid. Their protease resistance shows a pattern similar to that seen in extracts of [URE3] strains, with transiently stable 30 and 32 kDa species and a stable 7-10 kDa species that reacts on Western blots with antibody specific for the prion domain of Ure2p. Adding a small amount of cofilaments to an excess of native soluble Ure2p induces the conversion of most of the Ure2p to amyloid (66).
These results indicate that amyloid formation is the basis for the [URE3] phenomenon. Amyloid formation in vitro is promoted by the same part of the molecule (the prion domain) that is responsible for prion formation in vivo; the pattern of Ure2p protease resistance is similar for extracts of [URE3] strains and amyloid formed in vitro; Ure2p is aggregated specifically in [URE3] strains. It remains to be shown that Ure2p amyloid is in fact present in [URE3] strains.
Fig. 8. Amyloid fiber formation by native Ure2p directed by the Ure2p1-65 prion domain. Chemically synthesized Ure2p1-65 spontaneously forms 45 A diameter amyloid filaments in vitro (A) (66). Mixing equimolar amounts of synthetic Ure2p1-65 with native soluble full length Ure2p produces 200 A diameter cofilaments (B) which, on digestion with proteinase K, yields core-resistant filaments composed mostly of the prion domain of Ure2p (C) (66). Addition of small amounts of cofilaments (B), to excess soluble native full length Ure2p, results in seeding of polymerization of Ure2p forming 400 A amyloid filaments (D) (66).
Fig. 9. Birefringence of Ure2p amyloid filaments on staining with Congo red.
Role of Chaperones in Prion Propagation
One dramatic dividend from the discovery of yeast prions has been the finding by Chernoff et al. that chaperones have a critical impact on prion propagation (67,68). Heat shock protein 104 (Hsp104) is a chaperone of yeast that is capable of solubilizing proteins that have aggregated as a result of exposure to high temperature (69). It is the only heat-shock protein in yeast that is essential for surviving heat shock, and not for growth within the usual temperature range (70).
Overexpression of Hsp104 results in loss of [PSI+], accompanied by disappearance of the Sup35p aggregates (67,68). This effect is probably explained by the protein-disaggregating ability of Hsp104. Deletion of the HSP104 gene also results in loss of [PSI+] (68). Thus, Hsp104 levels must be optimal for [PSI] propagation. Overproduction must disaggregate the Sup35p amyloid, but why does underproduced Hsp104 result in loss of [PSI]? Two models have been proposed. One posits that Hsp104 is necessary for the generation of an intermediate form of Sup35p, which can then be converted to the [PSI+] form (68). Another model explains the requirement for Hsp104 by assuming that, without Hsp104, the aggregation would proceed to the extreme of forming just one lump in the cell, which would then be segregated to only one of the daughter cells; the other is, in effect, cured of [PSI+] (60). It is suggested that normal levels of Hsp104 break up the Sup35p amyloid into smaller lumps, thus assuring each daughter cell of receiving one or more fragments of amyloid. The fact that Sup35p amyloid formation proceeds well in vitro, in the absence of Hsp104 (63), argues against the first model, but further work will be needed to resolve this issue.
Hsp70s are a class of chaperones that assist protein folding and block aggregation or denaturation, by binding to hydrophobic patches on the surfaces of partially folded (or partially unfolded) proteins (reviewed in refs. 71 and 72). The curing of [PSI+] by overproduction of Hsp104 is partially blocked by the simultaneous overproduction of Ssa1p, one of the family of yeast Hsp70s (73). Another family of Hsp70 proteins, the Ssb family, functions in assisting the folding of newly synthesized proteins (74). The Ssb proteins appear to have effects opposite those of the Ssa proteins. Overproduction of Ssb proteins slightly increases, and elimination of Ssbs significantly decreases, the efficiency of curing of [PSI+] by overproduction of Hsp104 (75). Neither effect is mediated by an effect on the levels of the Hsp104 protein (75). These results reflect the complex interactions among heat shock proteins, and, in some cases, may be caused by the regulatory effects of one heat shock protein on others, either directly or by producing a state of stress in the cell. In any case, it is clear from the work of Chernoff et al. (75) that chaperones play central roles in the generation and propagation of prions.
Role of Interacting Proteins in [URE3] and [PSI] Generation
Recent evidence indicates that Mks1p is involved in the nitrogen regulation pathway, communicating the presence of ammonia in the medium to Ure2p and inhibiting Ure2p action on Gln3p (Fig. 2; 29). We find that deletion of MKS1 prevents the de novo generation of [URE3], even when Ure2p or one of its fragments is overproduced (Edskes and Wickner, in preparation). Although the mkslA strains are deficient in generation of new [URE3] elements, they are not defective in propagation of [URE3] introduced into the cell by cytoplasmic mixing. [URE3] in such cells is essentially as stable as in wild-type cells, and is similarly curable by guanidine. The inability to generate a new [URE3] is not the result of failure to overexpress Ure2p, nor is there evidence for an altered pattern of degradation of Ure2p. It will be of interest to determine whether Ure2p interacts directly with Mks1p. These results suggest that interaction of Ure2p with Mks1p makes it more suitable for conversion to the prion form (Edskes and Wickner, in preparation).
MKS1 was first identified as a gene negatively regulating growth, and is itself inhibited by the ^AS-cyclic adenosine monophosphate (cAMP) system (76). We found that a constitutively active allele of Ras2p also resulted in dramatically reduced generation of [URE3] on overproduction of Ure2p, consistent with the earlier results (Edskes and Wickner, in preparation). MKS1 was independently identified as LYS80, an inhibitor of lysine biosynthesis (77). It is not yet clear how this phenotype relates to the role of Mks1p in nitrogen regulation and prion generation.
These results indicate that cellular regulatory mechanisms can dramatically affect the de novo formation of a prion. With further understanding of the control pathways operating on Ure2p as part of the nitrogen regulation system, it is likely that other factors influencing prion generation will be found.
In normal cells, the Sup35 protein is known to be present as a soluble heterodimer with Sup45p, and this heterodimer is responsible for recognition of translation termination codons and release of the completed peptide. Derkatch et al. have found that overproduction of the Sup45 protein blocks the induction of the de novo formation of [PSI+] (78). This result indicates that it is the free form of Sup35p that is most capable of undergoing the prion change, and that, in its complex with Sup45p, it is stabilized (78).
The inhibition of prion formation by Sup35p on overproduction of Sup45p is the reverse of the inhibition of prion formation by elimination of Mks1p. The proteins interacting with HET-s or PrP are not yet known, but it is tempting to speculate that they too will affect prion generation.
[PIN+], A Non-Mendelian Genetic Element Controls Generation of [PSI+]
Derkatch et al. (79) have found that strains differ in their ability to give rise to [PSI+], even when Sup35p is overexpressed. Genetic analysis shows that the ability to give rise to [PSI+] in this way requires the presence of a dominant nonchromosomal genetic element called [PIN+] (for [PSI+] induction) (79). [PIN+] does not require the N-terminal part of Sup35p for its propagation, but whether it requires the C-terminal part of Sup35p cannot be tested.
Although overproduction of Hsp104 cures [PSI+] efficiently, it does not cure [PIN+] (79). In contrast, guanidine cures both [PSI+] and [PIN+] (79). This suggests that guanidine does not cure simply by inducing Hsp104 production.
Although [pin-] strains cannot be induced to generate [PSI+] by overproduction of intact Sup35p, they are fully capable of becoming [PSI+] by overproducing the Sup35p prion domain (79). Derkatch et al. (79) suspect that [PIN+] is a prion, but demonstration of this idea will be difficult, until a candidate protein is found whose overproduction induces [PIN+], and which is necessary for [PIN+] propagation.
![The Ure2 protein induces de novo [URE3] formation. Overproduction of Ure2p from an inducible GAL1 promoter results in an increased frequency with which [URE3] arises (7). Adding one nucleotide in codon 80 of URE2 produces a prion domain fragment with greatly elevated [URE3]-inducing activity (48). Further removing one nucle-otide in codon 44 changes the reading frame of most of the prion domain, eliminating [URE3]-inducing activity, without changing the level of mRNA (48,54). It was confirmed that the nucleotide removed from codon 44 was not of special importance, by removing the entire codon 44; this did not affect prion-inducing activity (49) . The Ure2 protein induces de novo [URE3] formation. Overproduction of Ure2p from an inducible GAL1 promoter results in an increased frequency with which [URE3] arises (7). Adding one nucleotide in codon 80 of URE2 produces a prion domain fragment with greatly elevated [URE3]-inducing activity (48). Further removing one nucle-otide in codon 44 changes the reading frame of most of the prion domain, eliminating [URE3]-inducing activity, without changing the level of mRNA (48,54). It was confirmed that the nucleotide removed from codon 44 was not of special importance, by removing the entire codon 44; this did not affect prion-inducing activity (49) .](http://what-when-how.com/wp-content/uploads/2011/08/tmpD64_thumb.jpg)
![Aggregation of Ure2p in [URE3] cells. A Ure2p-green fluorescent protein fusion protein, which was both active in nitrogen regulation and capable of involvement in the prion change, was expressed in wild-type, [URE3] prion-containing, and cured cells. In normal cells, the fusion protein was evenly distributed throughout the cytoplasm, but in prion-containing cells it was aggregated (65). Aggregation of Ure2p in [URE3] cells. A Ure2p-green fluorescent protein fusion protein, which was both active in nitrogen regulation and capable of involvement in the prion change, was expressed in wild-type, [URE3] prion-containing, and cured cells. In normal cells, the fusion protein was evenly distributed throughout the cytoplasm, but in prion-containing cells it was aggregated (65).](http://what-when-how.com/wp-content/uploads/2011/08/tmpD65_thumb_thumb.jpg)