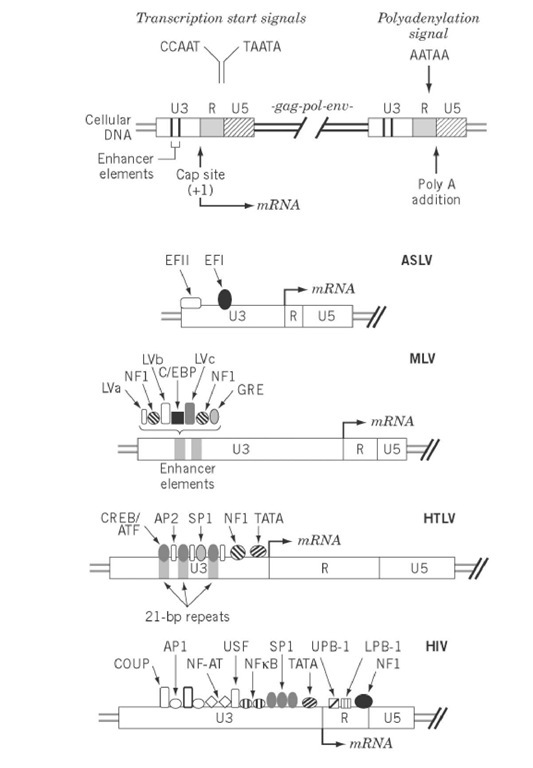
Within retroviruses, the integrated, double-stranded proviral DNA genome is flanked at its 5′- and 3′-termini by identical noncoding regions designated long terminal repeats (LTR). Each consists of three "domains." U3 and U5 are derived from unique sequences at the 3′ and 5′ termini of the viral RNA genome, respectively, and R denotes repeat sequences of the termini whose homology is exploited to transfer nascent DNA within or between genomes during proviral DNA synthesis. The domains are linked in the order -U3-R-U5-. Sites for initiation of minus- and plus-strand DNA (the primer binding site (PBS) and the polypurine tract (PPT)) are located immediately downstream and upstream of the 5′- and 3′-LTR, respectively. Although they provides many common control mechanisms, retroviral LTR vary considerably in size, ranging from as little as 350 bp in Rous sarcoma virus (RSV) to greater than 1700 bp in SFV-3 (Table 1). Size heterogeneity is also evident within individual LTR domains, for example, the R region of mouse mammary tumor virus (MMTV) has only 13 bp, whereas its human T-cell leukemia virus (HTLV) BLV counterpart has 235 bp. This contrasts with the U3 region of MMTV which, at almost 1200 bp, is among the largest reported. Currently, the reason for such size heterogeneity amongst retroviral LTR is unclear.
Table 1. Size Variation among U3, R, and U5 Components of Retroviral LTRa
a Most notable among these are the extremely short R elements, which play a pivotal role in DNA strand transfer during replication.
Despite their identity, 5- and 3′-retroviral LTR assume different roles during the expression of proviral DNA. U3 of the 5′-LTR harbors the promoter, a binding site for the host-coded RNA polymerase II and a variety of cellular protein factors that cumulatively govern the level of transcription. Viral RNA initiates at the first base of the 5′-R, and extends to the end of its 3′-counterpart, where it is polyadenylated . Additional levels of control have been identified within R and U5 sequences of the transcript, including (i), the HIV TAR loop, which interacts with virally coded transactivator proteins (eg, Tat) to augment transcription and (ii) U5-IR stem-loop structures, which interact through a variety of mechanisms with the tRNA replication primer to control initiation of minus-strand synthesis. Thus, the LTR can be considered a major contributor toward transcription and reverse transcription.
1. LTR control of viral transcription
1.1. The LTR Promoter
The LTR promoter provides the binding site for RNA polymerase II and accessory host cellular factors that interact with U3 sequences upstream from the TATA box, a consensus sequence 20 to 30 bp upstream of the transcription initiation site that mediates polymerase recognition. The complexity of cis-acting sequence elements in LTR promoters, shown in Fig. 1, varies from relatively few in the case of ALSV to as many as 10 in HIV-1. Several spatially distinct binding sites for a single accessory protein may exist (eg, AP-2 sites on the HTLV promoter) in addition to multiple adjacent binding sites (eg, the NF-AT and NFkB sites on the HIV promoter). Although these recognition elements are depicted primarily upstream of the transcriptional initiation site, precedents exist for regulatory elements more distal from the promoter. An example of this is the presence of a binding site for the C/EBP transcriptional factor in the gag gene of ALSV. The TATA box and upstream elements are commonly called the basal promoter.
Figure 1. Transcriptional control elements of retroviral LTR. The upper portion represents the integrated form of the provirus and indicates a variety of cis-acting elements in the U3 and R regions of the 5′- and 3′-LTR that regulates viral transcription. Binding sites for cellular transcriptional factors in the LTR of ALSV, MLV, HTLV, and HIV are indicated.
1.2. Enhancers
The enhancers, an important class of cis-acting elements are sequences that specify binding of cellular factors to enhance the basal level of transcription. Core enhancer sequences are relatively small (10 to 15 bp), but they collectively constitute a hierarchy of binding domains to generate one or more copies of an element functional independent of both position and orientation. Examples of this is are the tandem, 75-bp murine leukemia virus (MLV) enhancers depicted in Fig. 1, which sequester seven transcriptional factors.
Stimulation of transcription by viral enhancer elements has a variety of biological consequences. Although avian leukosis virus (ALV)-induced B-lymphomas lack an oncogene they reflect enhancer activity derived from proviral integration adjacent to the promoter for c-myc proto-oncogene. Proviral integration on either side of the proto-oncogene promoter can stimulate its transcription. In contrast, Rous-associated virus type 0 (RAV-0) infections are nonpathogenic, which correlates with lack of LTR enhancer activity in this virus. Deleting an enhancer element within the MLV LTR reduces the capacity to induce thymic lymphomas in mice.
Enhancer elements also play an important role in tissue-specific expression. Wild-type Moloney murine leukemia virus (MoMLV) is not effectively expressed in the liver or brain of mice. However, when the MoMLV LTR enhancer is replaced with its counterpart from the cellular transthyretin gene and a promoter-proximal sequence that controls tissue-specific expression, the resulting recombinant virus is infectious and is expressed in previously nonpermissive tissues. These observations are clinically significant because they open the possibility of constructing tissue-specific recombinant retroviral vectors for gene therapy.
1.3. Regulation of LTR Transcription
MMTV, a murine retrovirus responsible primarily for mammary tumors, provides a well-studied example of LTR expression regulated by glucocorticoid hormones. The glucocorticoid receptor contains an N-terminal activation domain, a central DNA binding motif, and a C-terminal ligand-binding site. After hormone treatment, receptor-bound hormone is transported to the nucleus, resulting in hypersensitivity of chromatin near glucocorticoid response elements (GRE). Accessibility of these exposed regions to transcription factors results in enhanced RNA polymerase II activity from promoters containing GRE motifs. The MMTV LTR contains multiple copies of the GRE. In the absence of hormone induction, a neighboring binding site in the LTR for the nuclear factor NF-1 is inaccessible because of association with nucleosomes . However, after induction, transport of the hormone-receptor complex to the nucleus essentially "clears" chromatin from the retroviral LTR, making the NF-1 binding site accessible to the transcriptional activator and stimulating viral transcription.
1.4. Transactivation
Transactivation is a feature of many complex retroviruses (spumaviruses, lentiviruses, and HTLV-related viruses) whereby, in addition to host factors, virally coded transactivators stimulate LTR transcription (1). Stimulation may occur through an interaction with DNA sequences in U3 (eg, the HTLV Tax protein and the bel-1 and taf gene products of human and simian foamy viruses, respectively), or through binding to a particular sequence near the 5′-end of the RNA transcript (eg, HIV Tat proteins). Tax-mediated stimulation of HTLV transcription is mediated via three 21-bp repeat elements in U3 that serve as recognition sites for cyclic AMP-responsive element binding protein (CREB), a member of the cAMP response family. A direct interaction of Tax with the 21-bp repeats has not been documented and spawns the theory that Tax exerts its function through protein-protein interactions to modulate CREB. Although the transactivators of foamy viruses act through similar mechanisms, their target sequences share little in common with those of Tax.
The target of the Tat transactivator in HIV and related lentiviruses is an ordered structure near the 5′ end of the viral RNA transcript, the transactivator response element or TAR (Fig. 2). Through an exhaustive series of investigations, the Tat target on TAR has been located at a small U-rich bulge in the stem. Different mechanisms have been proposed for the stimulatory effect of HIV Tat. One model suggests overriding transcriptional termination, based on observations that, in the absence of Tat, transcripts that initiate in the LTR pause after synthesizing 60 to 80 nucleotides. It is envisioned that TAR-bound Tat (and possibly other cellular factors) thereby interacting with and stabilize the host transcriptional machinery. Model in vitro systems show that an alternative hypothesis envisages Tat acting at the stage of transcriptional initiation through an interaction with the transcription factor SP-1. This may be a fortuitous consequence of the proximity of TAR to the site of mRNA initiation, hence the possibility that Tat action reflects a combination of these events also cannot be ruled out. Finally, the observation that Tat is released from infected cells and enters neighboring cells to modulate transcriptional activity suggests a further role of this protein in HIV pathogenesis.
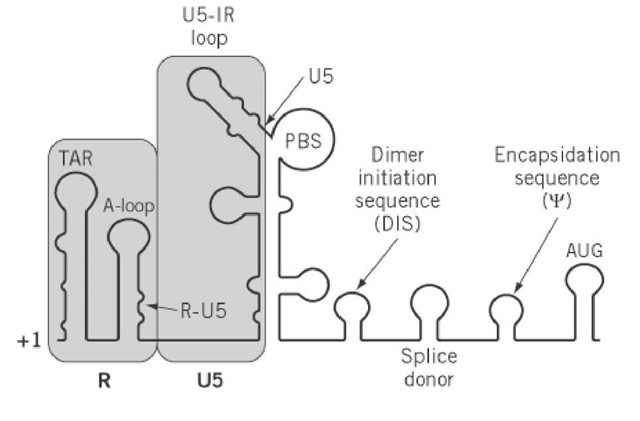
Figure 2. Schematic representation of secondary structural elements in the 5′-leader RNA of retroviruses that, it is proposed, regulate transcription and reverse transcription. +1 denotes the first nucleotide of the viral transcript, that is, the 5′-end of R. RNA that corresponds to the R and U5 regions of the LTR is shaded. Adjacent elements that control dimerization (DIS) and encapsidation of the retroviral RNA genome (Y) have been indicated. AUG represents the initiation codon of the gag open reading frame. PBS is the primer binding site.
2. R-U5 RNA and control of reverse transcription
The 5′-non-coding end of the viral transcript, that is, between the first nucleotide of R and the gag initiation codon, is defined as the leader RNA . Although the regulatory mechanisms differ among retroviruses, it is generally recognized that structural elements within the R-U5 portion of the leader play an important role in regulating the efficiency of reverse transcription. Extensive chemical and enzymatic probing has revealed a complex set of intramolecular stem-loop structures, as well as intermolecular duplexes involving different regions of the tRNA replication primer, the binding site for which lies immediately adjacent to U5 (Fig. 2). In discussing LTR control of reverse transcription, it is thus necessary to include RNA sequences that comprise the 5′-leader.
In addition to complementarity between viral PBS sequences and the 3′-terminal nucleotides of the tRNA replication primer, secondary intermolecular interactions critical to reverse transcription have been uncovered that involve U5 of RNA human and avian retroviruses. In ALSV, it has been elegantly demonstrated genetically and biochemically that bases comprising the TYC loop of tRNATrp (the cognate replication primer of ALSV; see Transfer RNA) interact with sequences in the U5 leader stem to control initiation of minus-strand DNA synthesis (2). Although secondary structural predictions for RNA folding suggest the potential for a similar mechanism in many retroviruses, HIV-1 adopts an alternative approach. In this case ( ), the U-rich tRNALys,3 anticodon domain is implicated in controlling reverse transcription, the target of which is the A-rich U5-IR loop in the immediate vicinity of the PBS (Fig. 2). Although both models are equally plausible, they must recognize a common requirement for disruption of RNA structural elements before and accompanying initiation of reverse transcription. For example, although hybridization of tRNATrp to the PBS of the ALSV genome has the consequence of unwinding its TYC stem, it is necessary to disrupt the U5 leader stem to provide the type of intermolecular duplex been proposed. Furthermore, once such non-PBS intermolecular duplexes are established, they must also be disrupted by the retroviral replicating machinery. The energy to disrupt such structures may be an intrinsic feature of RT, as proposed for the avian enzyme. Alternatively, this may be supplied by way of deoxynucleoside triphosphate hydrolysis during polymerization. Finally, accessory viral proteins, such as NC, could potentially interact with RT and serve a stimulatory role during initiation of reverse transcription.
Why are such complex regulatory mechanisms of reverse transcription necessary? One clue to this may lie in observations that alternative sites on the retroviral genome with considerable sequence homology to the PBS can be identified. Although initiation of reverse transcription from such "pseudo" initiation sites might be possible, this would have severe consequences at later steps in replication when the tRNA primer is removed from nascent minus-strand DNA. Minus-strand DNA sequences immediately adjacent to the tRNA primer are destined to become terminal nucleotides of the 3′-LTR critical for recognition by the retroviral integration machinery (see Fig. 4 of Retroviruses). Thus, although the PBS provides the appropriate sequence for localizing the tRNA 3′-terminus, additional tRNA/viral base pairing between could be envisaged as "locking" or stabilizing the replication primer at the appropriate initiation site. Support for this notion comes from genetically manipulated genomes of HIV-1, where the natural PBS is replaced by a variant that specifies binding of another tRNA isoacceptor species. If the PBS alone is exchanged, the resulting virus replicates poorly. In contrast, replication kinetics are significantly improved when sequences in the U5-IR loop are simultaneously altered to complement the anticodon loop of the heterologous tRNA isoacceptor.
In addition to the PBS, two non-LTR elements of the leader RNA should be pointed out. The first of these is the dimer initiation sequence (DIS), which promotes intermolecular base pairing of the RNA genome to ensure that a dimer of identical molecules is introduced into the budding virion. In HIV-1, palindromic sequences in the loop of a hairpin structure mediate dimerization through a mechanism defined as the "kissing complex." Finally, the encapsidation sequence (Y) provides a signal for packaging the retroviral genome into the budding virion that is mediated through an interaction with the NC component of the gag polyprotein. Location of Y between the major splice donor and the gag initiation codon ensures that spliced RNAs are not incorporated (see RNA Splicing). The exception to this is ALSV, whose splice donor is located in the gag gene, that is, downstream of the encapsidation sequence, and predicts that subgenomic RNA is transported to the virion. The observation that such species are in fact discriminated against suggests the existence of a cryptic encapsidation signal close to the 5′ end of the gag transcript.