The major histocompatibility complex (MHC) was first appreciated in mice as a set of proteins, encoded by closely linked genes on chromosome 17, that serve as the major targets for rejection of skin grafts. Humans were subsequently shown also to have MHC antigens, which are homologous to those found in the mouse but are encoded in the human leukocyte antigen (HLA) region on the short arm of chromosome 6 [see Figure 1]. Initially, human MHC antigens could be defined only by use of sera from multiparous women who had mounted humoral immune responses against the paternally derived MHC antigens in their fetuses. The development of DNA-based methods for genotyping of individuals has permitted more extensive study of these extraordinarily polymorphic molecules. This topic reviews the genetics and structure of the MHC, its function in the immune response, and its association with disease.
Structure and Antigens of the Major Histocompatibility Complex
There are two structural types of MHC molecules, called class I and class II. The molecules of both classes are active in antigen recognition and help focus immune defenses during invasions from the microbial world. They are also engaged in the communication that occurs between cells during the immune response. MHC molecules act by binding peptide fragments of antigens that have been processed in specialized antigen-presenting cells. Clonally determined antigen receptors on T cells then recognize and bind to specific peptide-MHC complexes, setting into motion the appropriate immune response. Segments of MHC molecules show sequence homologies with immunoglobulins, T cell antigen receptors, and T cell interaction molecules such as CD4 and CD8, which suggests that all these molecules share a common evolutionary ancestry.
The sequence and structure of MHC molecules have been extensively elucidated, and it has been determined that the polymorphic, or antigenic, portions of MHC molecules are quite small. In fact, the polymorphic portions frequently comprise only one to four amino acid substitutions encoded in regions of DNA nucleotide sequence hypervariability. A specific configuration in an MHC molecule resulting from particular substitutions of amino acids is called an epitope.
Mhc class i antigens
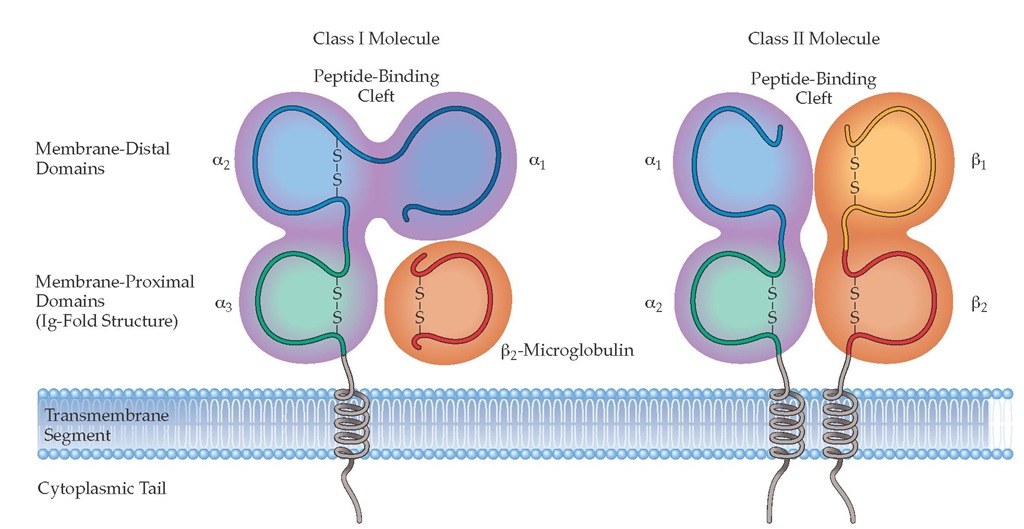
MHC class I antigens consist of two polypeptide chains held together noncovalently. One chain is heavy (44 kd) and glycosylated, and it determines antigen specificity. The extracellular portion of this class I heavy chain is divided into three domains, designated a1; a^ and a3. The other chain is a small (11.5 kd) protein known as ^-microglobulin [see Figure 2]. Class I heavy chains are the gene products of three MHC loci, designated HLA-A, HLA-B, and HLA-C [see Table 1]. There are many alleles for each locus; therefore, considerable polymorphism exists. ^-Microglobulin is encoded by a gene on chromosome 15. Both the ^-microglobulin and the a3 domain of the heavy glycosylated chain of MHC class I antigens demonstrate considerable structural similarity to the constant region of the heavy chain of IgG (CH3).
Figure 1 The best-characterized loci of the human major histocompatibility complex (MHC), located in the HLA region of the short arm of chromosome 6, are depicted. Distances are shown in recombination units (centimorgans), as determined by crossover frequencies in family studies, and in kilobases, as determined by sequence analysis of fragments produced by DNAses having defined cleavage sites. MHC class II molecules are encoded in the HLA-DP, HLA-DQ, and HLA-DR genes, and MHC class I molecules are encoded by HLA-B, HLA-C, and HLA-A genes. A cluster of closely linked complement genes—C4, BF, and C2—lies in the center of the region. There are two structural genes for C4, interspersed with two genes for the adrenal enzyme 21-hydroxylase. Next is the heat shock protein gene, Hsp70, followed by the tumor necrosis factor (TNF) genes, A and B. The orientation of the complement cluster and the TNF cluster has not been established, but an expanded view of this area could be depicted as -(C4-210HA-C4B-210HB-BF-C2)-(HSP70)-(TNFA-TNFB). GLO is a marker gene for the enzyme glyoxylase. An expansion of the class II region is in the lower portion of the figure. Each class II molecule is a heterodimer of an a and a p chain, which are encoded in the A and B genes, respectively. Pseudogenes, which are not expressed on the cell surface, are shown in white boxes. HLA-DP and HLA-DQ have one expressed heterodimer, A1B1; HLA-DR has only one A chain but nine genes for B chains (four are shown in the figure). The principal expressed heterodimers for HLA-DR are AB1, AB3, AB4, and AB5. In the region between HLA-DP and HLA-DQ lie the closely linked TAP1, TAP2, LMP2, and LMP7 genes. The TAP genes encode peptide transporters, whereas the LMP genes encode proteosomes that fragment proteins into peptides. This cytoplasmic system is believed to be responsible for production and delivery of peptides to MHC class I molecules before their movement to the cell surface.
MHC class I molecules have been crystallized, and their structure has been determined by x-ray diffraction to a resolution of 3.5 angstroms (A).1 Two of the heavy-chain domains, a1 and a2, are located at the membrane-distal portion of the heavy chain and form a groove along the top surface of the molecule. The sides of the groove are composed of a helices from the a1 and a2 domains, and the base is composed of eight antiparallel |-pleated sheets from these domains. The hypervariable (antigenic) regions are found mostly along the sides of the groove, but there is also variability in the | -pleated sheet region. The rest of the molecule shows minimal variability in relation to other molecules of the same HLA locus. In the crystals studied, the groove, which faces away from the cell membrane and is approximately 25 A long and 10 A wide, contains material representing processed antigen (i.e., peptide fragments). When peptides eluted from purified class I molecules are sequenced, they show patterns of amino acids, called motifs, that bind to particular sets of HLA class I molecules.2 These findings helped confirm the hypothesis that MHC molecules bind and present processed antigens to responding T cells and that the T cell receptor (TCR) recognizes foreign antigen as a peptide in the context of self-antigen; that is, it binds to a surface composed of both MHC and a bound peptide.
MHC class I antigens can be expressed on all cell types except erythrocytes and trophoblasts and can be detected by staining with labeled antibodies. Striated muscle cells and liver parenchymal cells are normally negative for class I antigens (i.e., they lack class I molecules or express only a low density of class I molecules), but in inflammatory states, these cells may become strongly positive for class I antigens.
Mhc class ii antigens
Some antibodies, elicited by immunizations with histoincom-patible cells, react with a limited variety of cells, most notably B cells, monocytes, dendritic cells, and activated T cells. Normally, these cells are the only ones found to bear MHC class II antigens. As is the case with class I antigens, however, inflammatory states cause many tissues to express class II antigens.
Each MHC class II antigen consists of two membrane-inserted glycosylated polypeptides, designated a (34 kd) and | (28 kd), which are bound together noncovalently [see Figure 2]. The extracellular portion of the a chain is divided into two domains, designated a1 and a2; the extracellular portion of the | chain is also divided into two domains, | 1 and | 2. Class II antigens are encoded by the HLA-D region, which is divided into at least three sub-regions: HLA-DP, HLA-DQ, and HLA-DR [see Figure 1].
Crystallographic studies indicate that MHC class II molecules have a structure similar to that of MHC class I molecules, with the a1 and | 1 domains forming a groove in which | -pleated sheets form the base and a helices form the sides.3 As in MHC class I molecules, the hypervariable (antigenic) regions of MHC class II molecules are located primarily along the groove, which again indicates a molecular basis for TCR recognition of foreign antigen together with self-MHC.
Class II MHC antigens can be identified by the use of sera from multiparous women that react predominantly with B cells. A serum is first exposed to platelets from a pool of many persons, because platelets contain MHC class I, but not MHC class II, antigens and thus will absorb antibodies to class I antigens, leaving antibodies to class II antigens in the serum. The naming of genes from the HLA-D region is now based on knowledge of the biochemistry of expressed antigens and on a growing database of DNA nucleotide sequences. The gene encoding the HLA-DR a chain, for example, is called DRA. Similarly, the closely linked genes encoding the | chains have been named DRB1 (encoding the | chains for DR1 through DR18), DRB3 (encoding the | chain for DR52), DRB4 (encoding the | chain for DR53), and DRB5 (encoding the | chain for DR51). Because DRB2 expresses no protein product, it is called a pseudogene. Each of the HLA-DR | chains associates with the common non-polymorphic HLA-DRA a chain to form functional class II HLA-DR molecules. HLA-DRA a chains are always the same; the difference in HLA-DR antigenic alleles is accounted for by variations in the genes encoding the HLA-DR | chains. The HLA-DQ locus contains the genes DQA1, DQB1, DQA2, and DQB2. DQA2 and DQB2 are pseudogenes, whereas the products of DQA1 and DQB1—that is, the a and | chains of HLA-DQ— are both polymorphic. HLA-DP gene organization is similar to that of HLA-DQ [see Figure 1 ].
Nomenclature of hla antigens
The nomenclature of the HLA system is coordinated through the World Health Organization Nomenclature Committee for Factors of the HLA System.4 The prefix for the gene name is HLA, followed by a hyphen, then a locus name (e.g., DRB1, DQA2, C).The alphanumeric identifier is composed of up to nine characters: AACCSSXXN, where AA is an integer that refers (when possible) to the serologic family of which the allele is a member; CC is an integer defining the nucle-otide coding variant resulting in a unique peptide product; SS is an integer defining synonymous variants (different DNA sequence but same amino acid sequence) of a coding variant; XX is an integer defining variants outside the coding region. An N is appended to the identifier if the allele is a null or nonexpressed variant. Thus, HLA-DRB1*030502 encodes a DR molecule that is serologi-cally in the DR3 group and has a different amino acid sequence from DRB1*0301, DRB1*0302, DRB1*0303, or DRB1*0304. It also has a different nucleotide sequence from DRB1*030501, but has the same amino acid sequence, making it a synonymous variant.
Frequency of different hla alleles
Two terms, haplotype and linkage disequilibrium, describe important associations between MHC genes. Haplotype refers to the set of closely linked genes on any one chromosome. Every person has two haplotypes of the MHC, one from each parent. Each haplotype has a particular set of antigens determined by the HLA-A, HLA-B, HLA-C, HLA-DR, and other loci.
The second term, linkage disequilibrium, refers to the observation that in a population, some HLA antigens coincide within a single haplotype much more frequently than expected. If discrete genes were distributed independently throughout the population, the frequency at which any two linked antigens encoded at different loci would occur within a haplotype is the product of their frequencies in the population. However, in whites, the HLA-A1 antigen and the HLA-B8 antigen are associated six to 21 times more often than would be predicted from their gene frequencies.
Figure 2 MHC molecules are of two structural types with very similar peptide-binding sites on the membrane-distal surface. (a) MHC class I molecules consist of heavy chains made up of three polypeptide domains (aj, a2, a3) and a noncovalently associated light chain, ^-microglobulin. (b) MHC class II molecules are heterodimers of a and p chains with a very similar overall structure and peptide-binding surface.
Such linkage disequilibrium may occur because not enough evolutionary time has elapsed for the genes governing the antigens to be evenly distributed or because such an association results in a selective advantage to the individual. Recombination, or crossover, takes place during meiosis and occurs about 1.5% of the time between MHC class I and MHC class II loci. Over many generations, recombination leads to an equilibrium of linked alleles in a population unless selective pressures favor survival of certain haplotypes. A hypothetical example of such selection would be the survival of persons bearing HLA haplotypes that confer resistance to epidemics, such as smallpox and plague. Racial differences are reflected in marked variations in the frequencies of certain HLA antigens and haplotypes. Fewer, though often striking, examples of such differences are also observed in various ethnic groups.5
Role of MHC in Immune Response
The mixed lymphocyte reaction
When lymphocytes from one person are cultured with those from another, the cells are stimulated to divide. This division, which can be measured from the rate of uptake of 3H-thymi-dine into the cells, is called the mixed lymphocyte reaction (MLR). By preventing the division of one of the sets of cells by treatment with mitomycin or irradiation, it is possible to study the antigens on the membrane of the treated cells that stimulate this proliferative response. In humans, HLA-DR antigenic determinants are mainly responsible for evoking a primary MLR. HLA-DQ antigens play a lesser role, and HLA-DP antigens do not appear to be involved in the primary MLR. However, responding lymphocytes that have been primed by previous exposure to HLA-DQ or HLA-DP antigens proliferate vigorously when reexposed to the same antigen in a secondary MLR. The primary MLR is driven by the very high precursor frequency of naive cells having affinity to HLA-DRB1, not by primed memory cells.
Antigen processing and presentation
The breakdown of protein molecules into peptide fragments is an important part of the process by which antigens are presented to T cells and other immune effector cells. MHC molecules come to the cell surface with peptides already bound. Proteins are first degraded internally, and the peptide fragments are bound to MHC class I and MHC class II molecules within the cell. Class I molecules are expressed on virtually all tissues. Virally infected cells are recognized principally by class I-restricted T cells, usually those with a cytotoxic function.
Table 1 Antigens of the HLA System
|
Antigen |
Number of Antigenic Specificities* |
Number of DNA Variantsf |
|
|
HLA-A |
22 |
290 |
|
|
Class I |
HLA-B |
48 |
555 |
|
HLA-C |
9 |
140 |
|
|
Class II |
HLA-DR (1-18) |
17 |
356 |
|
HLA-D |
24 (cellular) |
356 |
|
|
HLA-DR (51-53) |
3 |
76 |
|
|
HLA-DQ |
7 |
25 (DQA1) |
|
|
56 (DPB1) |
|||
|
HLA-DP |
6 (cellular) |
20 (DPA1) |
|
|
106 (DPB1) |
Antigenic specificities are defined by reactivity with HLA-specific sera from multiparous women (serologic specificity) or by proliferative response of cells (cellular specificity).
Variants at each locus; most variants result in a different expressed amino acid sequence.
Table 2 Cell-Mediated Lympholysis in a Mixed Culture
*The stimulator cells that induce proliferation of the responder T cells in the mixed lymphocyte culture reaction also serve as the targets for the cytotoxic cells that develop from the responder population (as measured by the cell-mediated lym-pholysis assay).
Low numbers of cytotoxic T cells may develop against class II antigens. ^Stimulator cells from two individuals are mixed with responder cells from a single individual.
In contrast, class II-directed T cells are restricted to antigen-presenting cells of the immune system (i.e., B cells, macrophages, dendritic cells, or Langerhans cells) that are principally concerned with defense against external infectious agents. Because class II-positive cells also carry class I molecules, they may act as antigen-presenting cells for both exogenous and endogenous proteins [see 6:IV Cell-Cell Interactions, Cytokines, and Chemokines in Immune Response Mechanisms].
Exogenous and endogenous antigens reach the cell surface by different pathways. Exogenous proteins are taken up into endo-somes or lysosomes, where they are catabolized. Peptides from exogenous proteins are generally bound to MHC class II molecules, and the class II-peptide complexes are then brought to the surface for presentation to T cells. Peptides from endogenous proteins (e.g., secretory proteins or products of viral infection) appear to be complexed in the endoplasmic reticulum to MHC class I molecules. Genes called LMP, which are also located in the MHC region, encode proteins that are responsible for breaking down proteins into small peptides (eight to 10 amino acids long); closely linked TAP genes encode chaperones that transport peptides across intracellular membranes [see Figure 1].6-8 This system delivers peptides of intracytoplasmic origin to newly formed class I molecules. As noted, certain peptide sequence motifs are known to be characteristic of peptides eluted from purified molecules of a given MHC allele.9,10 These findings indicate that the allelic sequence differences at the margins of the pep-tide-binding groove determine which peptide sequences will bind. Class I-bound peptides are usually nine amino acids long, with residues at particular locations that have similar charge or hydrophobicity (e.g., at positions 1, 3, and 9) for different groups of HLA alleles. In addition, a number of synthetic peptides representing immunogenic portions of infectious agents or other foreign proteins align on similar common motifs. Peptides eluted from purified HLA-DR class II molecules are variable in length, up to 25 residues, and have a minimal length of 13 to 14 amino acids. The motifs for DR1 represent a positively charged residue at position 1, a hydrogen bond donor at position 6, and hydrophobic residue at position 10.11 Prediction of binding affinity for a given HLA sequence is becoming common practice for development of peptide vaccines and studies of the specific immune response to protein antigens.