Biomedical Engineering Reference
In-Depth Information
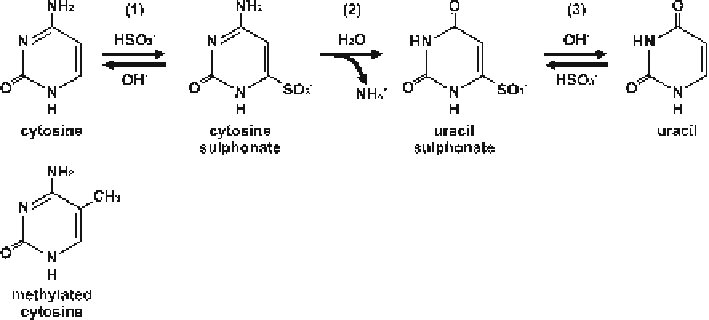
Fig. 10.3
Chemical reaction for the bisulfi te conversion of cytosine to uracil
since hydrolytic deamination caused by bisulfi te at the C6 position is blocked by the
presence of the methyl group at C5. Using next-generation-sequencing techniques
like the HiSeq2000 platform (Illumina), high-resolution genome-wide DNA meth-
ylation patterns with high coverage can be obtained. For local profi ling, bisulfi te-
specifi c PCRs can be performed using a strand-specifi c primer set detecting the
methylation information on one strand of the DNA. During PCR, 5-methylcytosine
residues are amplifi ed as cytosines and unmethylated cytosines are amplifi ed as
thymines (Fig.
10.4a
). The PCR products are cloned into proper plasmids and
sequenced using Sanger chain termination technology (Sanger et al.
1977
).
Comparison of the sequence of bisulfi te-treated DNA to genomic DNA allows the
identifi cation of 5-methylcytosine sites. To standardize the identifi cation of methyl-
ated cytosines within the amplicon, semiautomated tools are available (BiQ
Analyzer, Bock et al.
2005
). For local profi ling, next-generation-sequencing tech-
niques are also available by now using the 454 GS-FLX pyrosequencing platform
(Roche) with a freely available reasonable data evaluation pipeline (de Boni et al.
2011
; Lutsik et al.
2011
).
10.6.1
Bisulfi te Treatment of Honeybee Genomic DNA
Here we describe two approaches for bisulfi te treatment: (1) bisulfi te treatment of
genomic DNA packed in agarose beads and (2) bisulfi te treatment of genomic DNA
in solution. The fi rst method is suitable for the treatment of small amounts of total
genomic DNAs, while the second method is highly effi cient for converting cyto-
sines, but a loss of genomic DNA cannot be avoided. If enough genomic DNA is
available for repeating experiments, the second method is preferable.