Biomedical Engineering Reference
In-Depth Information
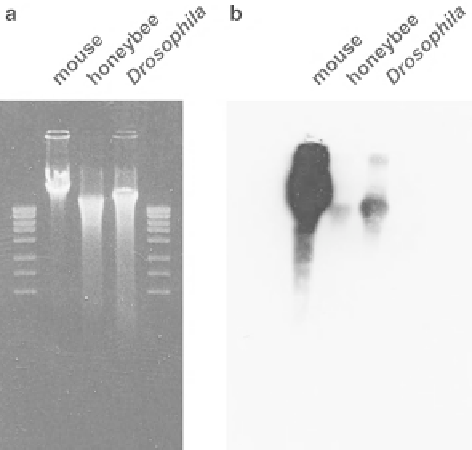
Fig. 10.2
Immunoblotting
with anti-5-methylcytosine
antibody. (
a
) Staining of the
agarose gel with ethidium
bromide showed that the
amount of genomic DNA
loaded from mouse,
honeybee, and
Drosophila
brains was similar. (
b
)
Although the honeybee
genomic DNA also had
methylated cytosines, the
amount in the total genomic
DNA was signifi cantly less
compared to the mouse and
Drosophila
genomes
patterns in genomic DNA. Enzymes such as
Hpa
II or
Hha
I only cut unmethylated
DNA leaving methylated DNA untouched. Subsequent Southern blotting provides
information about the relatively wide occurrence and abundance of 5-methylcytosine
in the genome (Rae and Steele
1979
; Quint and Cedar
1981
). This is a classical
method for investigating methylation patterns and recently used for invertebrates
(Krauss et al.
2009
; Robinson et al.
2011
). However, both immunoprecipitation and
methylation-sensitive restriction require a large amount of purifi ed genomic DNA
which is, especially when using tiny amounts of biological material, not always
available.
10.6
Bisulfi te PCR
Since more than a decade, bisulfi te conversion was developed for the sensitive iden-
tifi cation and direct mapping of the site of 5-methylcytosine using a very small
quantity of genomic DNA (Frommer et al.
1992
). This technique breaks down epi-
genetic information to the genetic level and is widely used for local, and nowadays
also genome-wide, methylation detection. The standard method for this technique is
based on the chemical conversion of unmethylated cytosines to uracils by treatment
of DNA with sodium bisulfi te (Fig.
10.3
). This chemical modifi cation precedes in
three steps: (1) sulfonation at the C6 position of the cytosine residue, (2) hydrolytic
deamination at the C4 position to produce uracil sulfonate, and (3) desulfonation
under alkaline conditions. The 5-methylcytosine remains unreactive to this process