Biomedical Engineering Reference
In-Depth Information
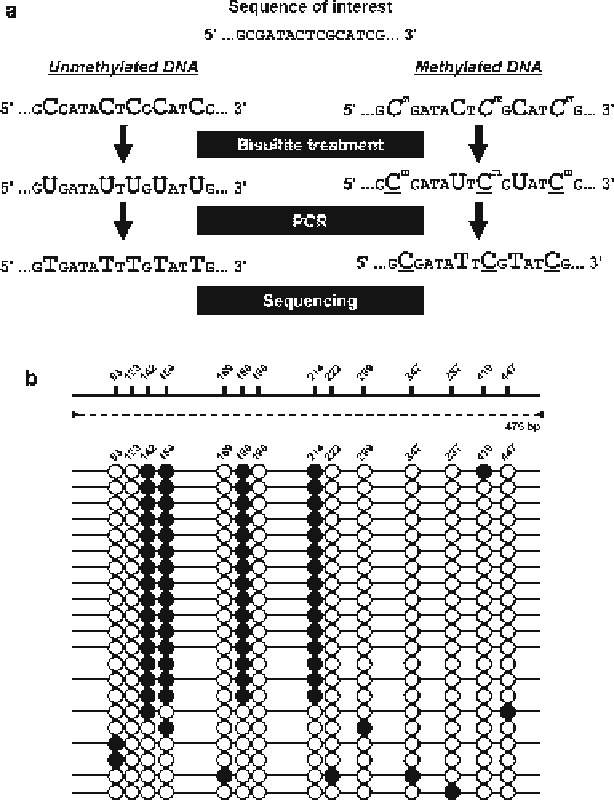
Fig. 10.4
(
a
) Principle of bisulfi te PCR. (
b
) Diagrams showing the methylation status of a
promoter region.
Filled boxes
on the
solid bars
indicate the candidate cytosine residues for meth-
ylation, and the number above each box indicates the nucleotide number counted from the fi rst
nucleotide of the PCR amplicon.
Arrowheads
and the
dotted line
represent primers and PCR
amplicon, respectively. Results using a primer set within the element showed a mixture of methyl-
ated (
fi lled circles
) and unmethylated (
open circles
) cytosines. Cytosines 142, 154, 188, and 214
were methylated in about 70 % of sequences
1 .
Bisulfi te treatment of genomic DNA packed in agarose beads
: Five hundred
nanogram of genomic DNA purifi ed from honeybee brains was treated over-
night with a restriction enzyme that does not cut within the target region to be
amplifi ed by PCR. Sodium bisulfi te powder (3.8 g; mixture of NaHSO
3
and
Na
2
S
2
O
5
; Sigma) was dissolved in 5 mL distilled water and 1.5 mL 2 M NaOH,
and 110 mg hydroquinone (Sigma) was dissolved in 1 mL distilled water and