Biomedical Engineering Reference
In-Depth Information
23
R
1
, R
2
=
H
24
coumarin
R
1
=
NH
2
H
H
NAA
1
AA
2
AA
3
NHR
2
O
HN
R
2
= fluoresceine
AA
1
-
3
=
Ala, Asp, Glu, Phe
Gly, His, Ile, Leu,
Met, Asn, Pro, Gln,
Ser, Thr, Val, Trp, Tyr
HN
H
O
AA
1
-
3
=
Ser-Tyr-Ser
NH
NH
N
AA
1
AA
2
AA
3
NHR
2
R
1
HN
H
2
N
H
17 × 17 × 17 = 4,913 receptors
Fig. 10 Receptor library 23 with 4,913 members and fluorescence labeled hit structure 24 for ATP
binding with amino acid sequence AA
1-3
¼
Ser-Tyr-Ser
interacts with the guanidinium moieties via hydrogen bond enforced salt bridges. For
ATP and ADP the nucleobase and the sugar moiety are inside the CyD cavity; for
AMP, on the other hand, only the nucleobase is included. This is probably due to the
distance between the cavity and the cationic template, which is too large to be bridged
by AMP's monophosphate hinge when sugar and nucleobase are inside the CyD.
1.4 Peptide-Based Receptors
As shown for carboxylate and oligopeptide recognition (vide supra), peptides have
proven to be excellent building blocks for the synthesis of artificial receptors.
Furthermore, they are excellent model systems for proteins to investigate, e.g.,
ATP binding sites, which are commonly present in many enzymes.
Anslyn used the combinatorial library 23 (R
1
,R
2
¼
H), shown in Fig.
10
, with
4,913 members (17
3
) to obtain ATP selective receptors [
15
]. Two combinatorial
tripeptide side chains were linked by an aromatic scaffold which preorganizes
adjacent groups alternately “up” and “down” with respect to the benzene ring. As
a result, a cavity is formed for substrate binding. Each arm was attached to the resin
via a guanidinium group in order to act as a phosphate binding site. The third arm
was used as the connection to the solid phase and features a lysine group. The
library was screened in buffered water at pH 7.1, with fluorescence labeled ATP,
revealing that approximately 15% of the library members bind to ATP. Several hit
structures were resynthesized and labeled with two different dyes: fluoresceine at
the N-termini of the two arms (R
2
) and coumarin in the third arm (R
1
, as internal
standard). Binding studies in solution verified the binding properties in all but one
example. This result again stresses the importance of confirming on-bead results
with those obtained in solution. The best receptor, with the amino acid sequence
AA
1-3
10
3
M
1
(pH 7.1, buffered
water). The competing analytes AMP and GTP were bound significantly less
effectively. The presence of Tyr and Ser in the best receptor hints at hydrogen
bonding and
¼
Ser-Tyr-Ser (24), binds to ATP with
K
¼
3
-stacking between receptor and adenine and/or ribose. By attaching
the fluorescence labeled receptors onto a solid support they could be used as
selective sensors in arrays [
16
].
p













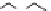





























Search WWH ::

Custom Search