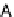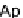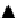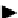Biology Reference
In-Depth Information
(a) Phosphorylation
(b) Specific activation of helicases
Fig. 6. PN modules, modeling enzymatic activities found in protein complex assembly. (a) The model of protein activation where
phosphorylation of a specific domain mediates interacting capabilities, thus, influencing subsequent complex rearrangements;
(b) a specific factor binds to a generic helicase, thus, mediating substrate specificity. (Colours are visible in the online version
of the article at
www.iospress.nl
.)
Signal reactions of the spliceosome assembly pathway
In an iterative process, the literature was inspected to isolate reactions involved in spliceosome as-
sembly. Reactions and involved proteins are based on human spliceosomal processes. All reactions are
ordered according to the four major stages of spliceosome assembly, E-, A-, B- and C-complex (see
Supplementary Table S1). After drafting each major stage the structural composition of the assembly
network was evaluated by T-invariant analysis. The net was only extended if each reaction is part of at
least one T-invariant. Reactions are only included if the factors could be integrated in a causal order.
As consequence, some spliceosomal factors,
e.g.
, SPF27, CypE, HSP73 and others, are omitted from
the model due to lack of evidence for a specific time point at which they participate in the spliceosomal
assembly process. Further, it is assumed that all factors for which no evidence of leaving or destabi-
lization in spliceosome assembly is given, implicitly remain in the model, until, finally a dissociation
into substructures (
e.g.
, snRNPs) and their recycling takes place. Hence, not all factors modeled with an
input transition have an explicit output transition. This is reasonable since many substructures remain
intact for repeated rounds of spliceosomal assembly [Pandit
et al.
, 2006]. This results in a model with
140 reacting species (places) comprised of RNA, proteins and intermediate complexes was established,
which covers about half of the currently known spliceosomal proteins (Fig. 7). The final network consists
of 161 transitions. 92 (57%) of them are boundary transitions, with 68 (74%) input and 24 (26%) output
transitions. In total, 69 (43%) transitions describe the internal reactions of the assembly network. This
biological network was modeled by reactions, which reflect a certain hierarchy that is characteristic for
spliceosome assembly. Hence, it can be seen as a process, in which almost all events are part of a causal
relationship, thus we expect the model not to reflect concurrent events, that are,
e.g.
, signaling pathways
that occur independent from each other.