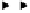Biology Reference
In-Depth Information
50 kb
cen
tel
Figure 4.26.
Map of the human type-I interferon gene cluster on chromosome 9p22 (after
Diaz
et al
., 1994). Solid rectangles: genes; open rectangles: pseudogenes.
and divergence (Miyata
et al
., 1985). The proximal genes,
IFNA1
,
IFNA2
,
IFNA5
,
IFNA6
,
IFNA13
, appear more closely related to each other than they are to the dis-
tal genes,
IFNA4
,
IFNA7
,
IFNA10
,
IFNA16
,
IFNA17,
and
IFNA21
, whilst
IFNA8
is equally divergent from both groups (Gillespie and Carter, 1983; Henco
et al
.,
1985; Miyata and Hayashida, 1982). Since these two groups are located at opposite
ends of the gene cluster, this may reflect an early multi-gene duplication. The gene
cluster appears to have evolved by gene duplication as a result of unequal crossing
over involving tandem units of
IFNA
and
IFNW1
genes (Diaz
et al
., 1994) but
divergence between gene sequences as well as subsequent gene deletions and dupli-
cations have served to erase the evolutionary history of parts of the gene cluster
(Diaz, 1995). The
IFNA14
gene, located midway between the two groups, is simi-
lar to the distal group in its 5
half (Diaz
et al
.,
1994). This is explicable either in terms of a unequal crossing over event between
distal and proximal interferon genes or a gene conversion event which has cor-
rected the 5
half and the proximal group in its 3
half of the gene against a donor proximal gene. Several of the human
IFNA
genes differ from the other family members by one or more relatively subtle
mutations. Golding and Glickman have shown in their insightful and prescient
(1985) study that these changes are explicable in terms of their templation by the
local DNA sequence environment. Thus, the in-frame deletion of a GAT codon in
the
IFNA2
gene may have been templated by a 5 bp inverted repeat 9 bp 5
to the
observed deletion (
Figure 4.27a
). Similarly, a 9 bp inverted repeat (separated by 25
bp DNA) may have templated an AA to GT change in the 5
flanking region of the
interferon “
9” gene (
Figure 4.27b
) whilst a 16 bp direct repeat could have tem-
plated five distinct sequence changes (two transitions, two transversions and a gua-
nine insertion) as the product of one mutational event (
Figure 4.27c
).
The type I interferon genes not only lack introns but also exhibit sequence
homology indicating that they share a common ancestry (Miyata and Hayashida,
1982). Since the spacer regions between primate
IFNA
genes still retain some
sequence similarity, we may surmise that some of the gene duplication events
have been relatively recent (Ullrich
et al
., 1982). Indeed, Miyata and Hayashida
(1982) estimated that they arose within the last 26 Myrs. The duplicational expan-
sion of the
IFNA
gene cluster may have been driven by the need to produce large
quantities of interferon rapidly, by selection for a novel temporal or spatial pattern
of expression, or by selection for a specialized novel function, perhaps associated
with a variant receptor with lower affinity for its ligand (Diaz, 1995).