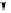Biology Reference
In-Depth Information
Insulin and insulin-like growth factor genes.
The insulin gene family has
ancient origins among the primitive eukaryotes (Le Roith
et al.
, 1980; Smit
et al.
,
1993). Human representatives of this superfamily include insulin (
INS
; 11p15.5),
insulin receptor (
INSR
; 19p13), insulin-like growth factor I (
IGF1
; 12q22-q24),
insulin-like growth factor II (
IGF2
; 11p15.5) and the relaxins (
RLN1
and
RLN2
;
9pter-q12). The origin of the
IGF1
and
IGF2
genes, which encode important reg-
ulators of growth and development, antedates the emergence of the first verte-
brates, since they appeared early on in the evolution of the protochordates more
than 600 Myrs ago (Chan
et al.
, 1990; McRory and Sherwood, 1997; Nagamatsu
et
al.
, 1991; Upton
et al.
, 1997). It is likely that in primitive organisms, insulin-like
peptides functioned so as to promote the uptake and utilization of nutrients, and
as a consequence, stimulated growth. With increasing complexity, nutrition
and growth became uncoupled and the insulin family diversified into proteins
and receptors with differing capacities for regulating metabolism and growth
(Steiner
et al
., 1985).
Interferon genes.
The interferon superfamily of viral defence proteins comprises
two main classes of gene (type I and type II). The type II interferons have only one
member, interferon-
, encoded by a gene (
IFNG
) on chromosome 12q14. By con-
trast, type I interferons comprise several sub-families of genes most of which are
clustered in a 400 kb region of chromosome 9p21. This cluster contains 13 inter-
feron-
genes (
IFNA1, IFNA2, IFNA4, IFNA5, IFNA6, IFNA7, IFNA8,
IFNA10, IFNA13, IFNA14, IFNA16, IFNA17
and
IFNA21
), a single interferon-
gene (
IFNB1
) and several pseudogenes
(Diaz
et al.
, 1994;
Figure 4.25
). The genes are arranged in tandem and most of the
functional genes are oriented with their 3
gene (
IFNW1
), a single interferon-
ends pointing in a telomeric direction.
There are two exceptions,
IFNA1
and
IFNA8
, which together with four pseudo-
genes orient towards the centromere (
Figure 4.26
). This is consistent with the
occurrence at some stage of an inverted duplication within the gene cluster with
its breakpoint between
IFNP12
and
IFNP11.
The evolutionary relationships of the various
IFNA
family members have been
determined by phylogenetic analysis (Golding and Glickman, 1985) and are con-
sistent with the emergence of this gene family by a process of gene duplication
A
D
C
B
A
D
CB
45S
A
B
18S
5.8S
28S
Figure 4.25.
Organization of the human ribosomal gene clusters (after Erickson and
Schmickel, 1985). The repeat pattern consists of four
EcoR
I fragments: A (7.3 kb), B (6.1
kb), C (11.7 kb), and D (16-19.6 kb) which together comprise a total repeat length of
41.1-44.7 kb. The inverted triangle denotes an area of length variability.
EcoR
I fragments
are indicated by arrows. The 45S precursor rRNA transcript is processed to yield the
mature 18S, 5.8S, and 28S rRNAs.