Chemistry Reference
In-Depth Information
N
S
-
S
O
hAGT
NH
hAGT
N
N
target protein
target protein
O
H
2
N
HN
H
2
N
N
probe
N
probe
H
scheme 2.25
Protein labelling via hAgT fusion protein.
O
R
Cl
O
NH
ASP106
O
N
O
R
CO
2
H
Luc8
O
HO
2
C
O
-
O
CO
2
H
ASP106
Halo
Q dot
HO
2
C
CO
2
H
Luc8
+
HO
2
C
CO
2
H
O
Q dot
Halo
HO
2
C
HO
2
C
HO
2
C
CO
2
H
Coelenterazine
CO
2
H
CO
2
H
O
655 nm
N
CO
2
H
R=
O
O
O
BRET
O
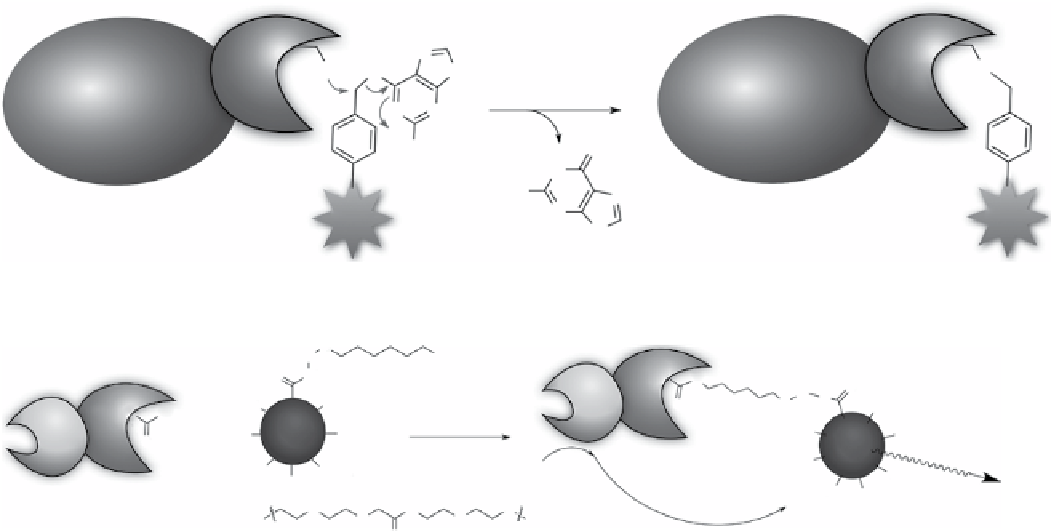
scheme 2.26
HaloTag used to conjugate luciferase and quantum dots.
with haloalkane dehalogenases (HaloTag, around 33 kda) can therefore be modified with different functionalities, such as
fluorophores, affinity handles, and solid support, selectively using synthetic ligands that bear haloalkane substrates. Los et
al. designed a modular system for protein labelling using HaloTag, which allowed modification of proteins in buffers and in
fixed or living cells and applied the protocol in imaging of subcellular protein (nF-κb proteins) translocation and protein-
protein or protein-dnA interactions [337]. Visualisation of cell membrane‒bound β-integrin molecules was achieved using
HaloTag fusion [338]. The HaloTag catalysed labelling occurs very rapidly in buffer, and a second-order rate constant was
obtained to be 2.7 × 10
6
M
-1
s
-1
, in the same speed scale as the binding between streptavidin and biotin (8.5 × 10
6
M
-1
s
-1
) using
nanomolar protein and ligand. In living cells, sufficient labelling was obtained at micromolar of ligand within 15 min.
using the HaloTag labelling protocol, the Rao group constructed quantum dot-luciferase conjugates using luciferase-
HaloTag fusion protein and quantum dots modified with haloalkane chains [339]. Quantum dots can also be fixed to a living
cell surface when the HaloTag is introduced to cell membrane bound protein genetically [340]. The Moerner group and col-
laborators demonstrated localisation of α-tubulin in cells transfected to express HaloTag-α-tubulin [341]. After addition of the
HaloTag ligand, a chloroalkylated photoactivatable dye dCdHF, cells were visualised using single-molecule super-resolution
imaging. Protein localisation in live
Caulobacter crescentus
bacteria was also visualised using transgenic organisms express-
ing HaloTag fusion proteins. The utility of molecular imaging using HaloTag in cells has also been shown in the imaging of
peroxisome dynamics in various cultured mammalian cells using C-terminal peroxisomal matrix protein targeting signalling
protein fused to HaloTag [342]. Real-time single-molecule imaging of
trans
-translation entry process was realised by anchoring
ribosomal protein to solid surface via HaloTag labelling [343].
In tumour models intraperitoneally implanted with cancer cells with highly expressed HaloTags,
in vivo
spectral fluores-
cence imaging of the tumour cells was obtained via endoscopy after administration of various green to near IR fluorophore-
conjugated HaloTag ligands, demonstrating a valuable tool for cancer research in animals [344]. In other work, tumours with
a high expression of α subunit hypoxia-inducible factor (HIFα) were visualised in live animals, which relied on the
construction of a near-infrared fluorescently labelled HIFα-targeted fusion protein via HaloTag labelling [345]. Recently,
doTA(gd) was introduced to protein via HaloTag labelling, and increased relaxivity (
r
1
and T
1
-weighted images) was
observed compared to unbound doTA(gd) [346]. The best doTA(gd)-protein conjugate exhibited sixfold enhancement in
r
1
compared to its unbound state. The increased relaxivity is probably caused by the receptor-induced magnetisation enhance-
ment effect while the motion of a contrast agent is coupled to that of a target protein.
eDHFR tag
utilising the tight and specific binding between
E. coli
dihydrofolate reductase (edHFR) and trimpethoprim
(TMP) (K
d
< 1 nM) [347], the Cornish and Sheetz groups developed a robust protein labelling method using an edHFR tag
(Scheme 2.27) [348]. Protein fused with edHFR could be labelled with near stoichiometric concentration of TMP-modified