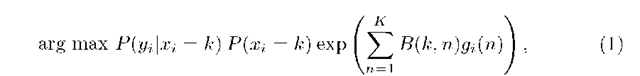Abstract
Organ localization is an important topic in medical imaging in aid of cancer treatment and diagnosis. An example are the pharmacokinetic model calibration methods based on a reference tissue, where a pectoral muscle delineation in breast MRI is needed to detect malignancy signs. Atlas-based segmentation has been proven to be powerful in brain MRI. This is the first attempt to apply an atlas-based approach to segment breast in T1 weighted MR images. The atlas consists of 5 structures (fatty and dense tissues, heart, lungs and pectoral muscle). It has been used in a Bayesian segmentation framework to delineate the mentioned structures. Global and local registration have been compared, where global registration showed the best results in terms of accuracy and speed. Overall, a Dice Similarity Coefficient value of 0.8 has been obtained which shows the validity of our approach to Breast MRI segmentation.
Keywords: breast MRI segmentation, probabilistic atlas, Bayesian framework, Markov Random Field.
Introduction
Atlas-based segmentation is a powerful generic technique for automatic delineation of objects in volumetric images that can take into account neighbourhood relationships between several different structures. Many atlas-based segmentation methods have been proposed in the literature for 3D medical imaging applications especially applied to segment MR brain images ([1,14] and references therein). However, a variety of these algorithms have also been used to different 3D image modalities and different body structures such as prostate MRI [5,8], abdominal CT [9], chest CT [10] and head and neck CT [3].
Neither previous work in atlas-based segmentation for the delineation of breast structures nor public breast MRI probabilistic atlases are present in the literature. In this work we have developed a multi-class probabilistic atlas-based segmentation method for breast MRI. The method segments the pectoral muscle, fatty and dense breast tissues, the heart and lungs. Segmentation of these structures is useful for cancer diagnosis or treatment applications where organ localization is needed. More specifically, our proposal aims to segment the pectoral muscle to use it as a reference tissue in pharmacokinetic model calibration [7,15]. Note that in this work a "probabilistic atlas" refers to the pair of an anatomical image and a probability tissue distribution volume. The former defines the atlas reference space, while the latter provides a volume with the complete spatial distribution of probabilities that a voxel belongs to one or more organs. Next sections describe the material employed (Sec. 2), the corrections applied to the volumes (Sec. 3.1), how the probabilistic atlas was built (Sec. 3.2) and how it is incorporated in a Bayesian voxel classification framework providing very rich information to find the organ location (Sec. 3.3). Qualitative and quantitative results are shown in Sec. 4 and finally conclusions are given discussed in Sec. 5.
Material
The data set used to construct the atlas and evaluate the segmentation results consists of 9 breast T1 weighted MR scans obtained from clinical data. Breast MRI examinations were performed on a 1.5 T system (Siemens 1.5T, Magnetom Vision), with a dedicated breast coil (CP Breast Array, Siemens, Erlangen). A dynamic contrast enhanced T1-weighted Flash-3D sequence was used, with repetition time of 8.1 ms, an echo time of 4 ms, and a flip angle of 20 degrees. The pixel spacing was 1.25 mm x 1.25 mm, and the slice thickness 1.5 mm. Per series, 108 slices were acquired, without interslice gap. Patients were scanned in prone position.
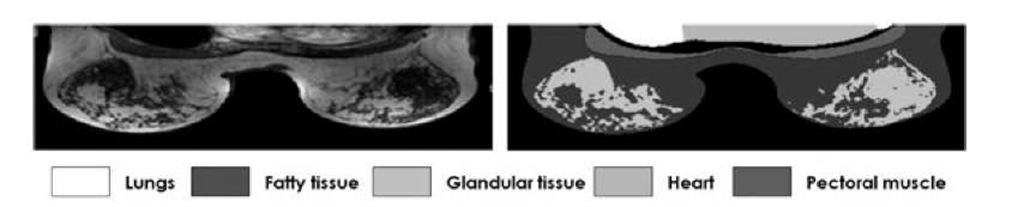
The pre-contrast series were used for the segmentation and each MR volume was manually segmented by an expert into 7 classes: background, fatty tissue, glandular tissue, pectoral muscle, lung area and the heart. The seventh class is the "non-of-above" class. Fig. 1 shows an example of a MRI slice on an axial view and the manual delineation of the mentioned classes.
Fig. 1. MR scan on an axial slice of a clinical breast MR T1 weighted volume with the manual annotation of the different structures
Methodology
This section describes the parts that compose the methodology. Firstly, a brief description of the preprocessing algorithms applied to the data set is given in Sec. 3.1. Secondly, Sec. 3.2 describes the method developed for building a breast probabilistic atlas. Then, adopting the classification made by Rohlfing et al. [11], Sec. 3.3 shows the Average Shape Atlas segmentation approach implemented in this work, which uses atlas information on a Bayesian framework.
Data Preprocessing
Because of the inhomogeneity of the breast coil sensitivity, intensity values are corrupted. Signal intensity homogeneity is required because image artifacts can considerably affect registration and segmentation results. For this reason the first step of the methodology consisted in correcting variability between images and inhomogeneities. Hence, image normalization algorithm was developed and applied to each scan in order to compensate inter-patient signal intensity variability. In addition, Non-parametric intensity Non-uniformity Normalization (N3) [13] bias field correction method was also employed to each scan.
Construction of the Atlas
In this work, for each patient segmentation, a full probabilistic atlas was built with the 8 remaining patients following a leave-one-out evaluation strategy. Firstly, the 8 patients and their segmentations were mapped onto the same reference space and the probabilistic atlas was created computing the frequency with which each location was labelled as a specific organ. A common reference space was used for all tests by selecting an extra patient that was not included in the evaluation data set. This extra image became the anatomical image of the atlas. Secondly, the final smooth probabilistic atlas was obtained using a 3D Gaussian convolution, basically to compensate for the small number of cases and the local registration errors. Variance a2 of 50 and 20 mm were used for global and local registration respectively. Fig. 2 shows an example of a pectoral probabilistic atlas before (a) and after (b) smoothing.
Fig. 2. Pectoral probabilistic atlas example before (a) and after (b) smoothing
For the first registration stage we employed two warping transforms to compensate for inter-patient differences. The first transform we evaluated was affine registration focused on a Volume of Interest (VOI). Although we were aware that such transform does not offer enough degrees of freedom (DOF) to compensate for the large differences that are present in this type of images (see Fig. 3), we observed that we could globally align pectoral muscles (which was the main goal of the segmentation) by aligning thoracic areas. For this reason, a VOI in each volume was defined by manually selecting the thoracic area with a single annotation point. Such approach was selected having in mind to obtain a good execution time to make the method suitable for clinical use.
The second evaluated transform was a non-rigid registration based on B-Splines proposed by Rueckert et al. [12]. This algorithm has been vastly applied by atlas-based segmentation methods [1,5,10,11] to minimize inter-individual variability in the shapes of anatomical structures and it is usually initialized by a global registration method like the affine registration described earlier.
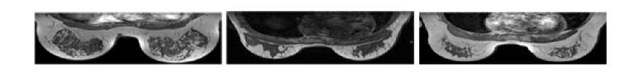
Fig. 3. Three breast MRI axial slices from three different subjects: variation between structures is easily observed in these three examples
Both approaches maximize the similarity measure of Mutual Information (MI) in an multi-resolution scheme using a stochastic gradient descent optimizer. For non-rigid registration, B-spline grid spacing of 32, 16, 8, 4 and 2 mm was used for each of the 5 resolutions respectively. Elastix [6] was used for the implementation.
Segmentation
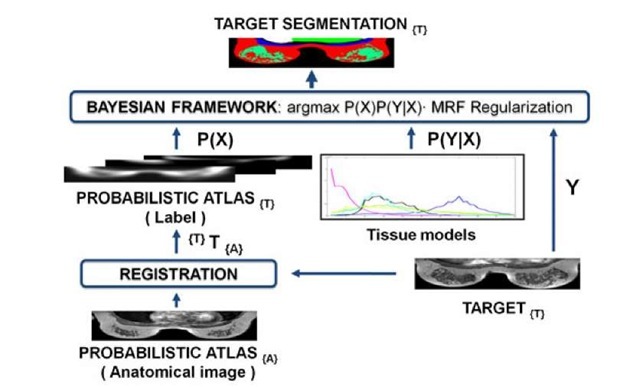
As we mentioned previously, the atlas is used in a Bayesian framework in order to segment the different structures of the breast. The approach is based on the work of Park et al. [9] and Karssemeijer [4], who segmented abdominal structures in computed tomography (CT) and three-dimensional X-ray images respectively. Figure 4 shows a schema of the Bayesian voxel classification algorithm incorporating the use of the probabilistic atlas.
Fig. 4. Voxel classification algorithm overview: from bottom to top, the labels of the probabilistic atlas are mapped onto target image space {T} using the anatomical image of the atlas. The probabilistic atlas, the tissue models and the target are provided to the Bayesian framework as a prior probability P(X), conditional probability P(Y|X) and set of intensity values Y, respectively. The Bayesian framework estimates the segmentation X that maximizes P(X)P(YIX).
In this paper the true label (the segmentation or set of labels) is denoted by X and the target image (data set of intensity values) is denoted by Y. Elements of X and Y are arranged by a spatial position denoted by i G I, where I is the simple index (x, y, z) in a 3D rectangular grid. Sample realizations of X and Y are represented throughout this work as x = (xi,x2, …,xN) and y = (yi,y2, …,yN), respectively, where N is the total number of voxels. Sample space of X is denoted nx where nx = {x : x, G {1, 2, 3, 4, 5, 6, 7}, Vi G I}. Labels 1, 2, 3, 4, 5, 6 and 7 are background, fatty tissue, glandular or dense tissue, heart, lungs, pectoral muscle and "None of the above" label respectively.
The problem consists in estimating the label X that best explains the given observation Y according to some cost function. As a decision rule, MAP (maximum a posteriori) was chosen: segmentation X was estimated by maximizing the global a posteriori probability P(X\Y) by searching the most probable labeling given the image Y and some prior model. Using Bayes theorem, the posterior probability to be maximized can be written as P(Y\X)P(X). The probability distribution P(Y\X) of the image Y, given a particular segmentation X, can be estimated from training data or from the image at hand. For example, Park et al. modelled probabilities of y^s as conditional Gaussians given mean and variance of the true label X. In this work P(Y\X) was specified by signal intensity tissue models directly built from the scans and manual segmentations of the data set. For each structure, a histogram of intensity values was built considering the voxels of the MRI volumes which belong to it using the manual segmentations.
On the other hand, the probability distribution P(X) is given by the probabilistic atlas once it has been mapped onto the target space using the same registration procedure used in its construction. A Markov Random Field (MRF) regularization is included to smooth the segmentation taking into account neighbourhood information. It introduces the probability of finding a particular label at i that depends only on the labels of voxels close to i (26 nearest neighbours in this work). Considering the addition of the MRF regularization, the posteriori probability, optimized by Iterated Conditional Mode (ICM), is defined as follows
where gi(n) denotes the number of neighbours labelled as n, K is the number of classes and the interaction parameters B(k, n) determines if regions labelled k and n are likely to neighbour each other.
Results
In a leave-one-out experiment we evaluated both registration methods by comparing the segmentation results. The quality of the segmentation was measured by determining the similarity with the ground truth. As a performance measure the Dice Similarity Coefficient (DSC) was calculated. Figure 5 shows a box plot with DSC values for the patient segmentations of each organ using affine registration (A) or B-Splines (B).
Fig. 5. Box plot with segmentation DSC values for each organ using VOI affine (A) and B-splines (B) registrations
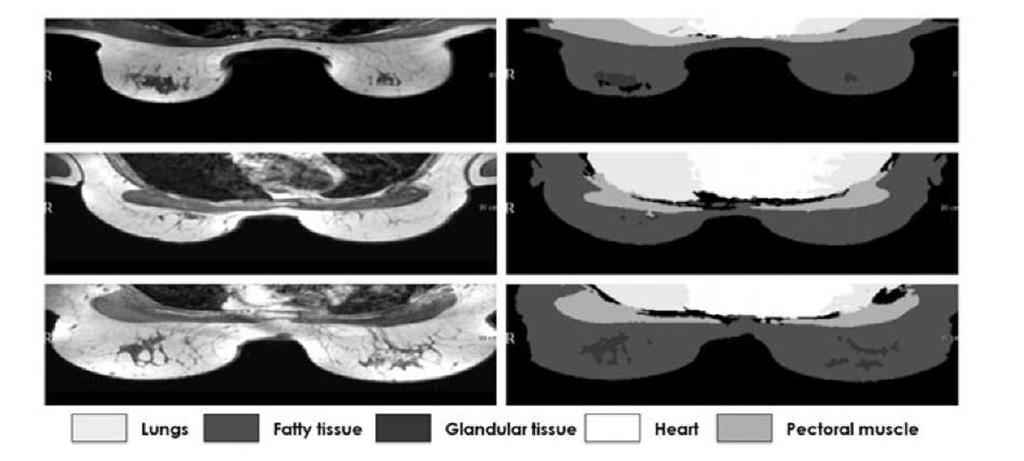
Fig. 6. Intermediate slices from 3 different patients and their segmentation using affine registration with VOI
For lungs and heart, VOI affine registration clearly outperforms B-Splines because non-rigid transform seems to introduce errors (p-value < 0.05, two-sided paired t-test). Segmentations of fatty and dense tissues and pectoral muscle provides similar values with no significant differences for both registration methods (p-values of 0.66, 0.43 and 0.15 respectively for a two-sided paired t-test). To sum up, mean DSC values were 0.8 for affine registration and 0.72 for B-Splines. The execution times of affine and B-Splines, measured on Intel(R) Core(TM)2 Quad CPU Q9550 2.83GHz were 15 min. to 1 hour aprox. respectively.
Overall, segmentation results can be considered satisfactory in all cases (normally a DSC > 0.7 is considered good segmentation [2]), which is illustrated by Fig. 6, where intermediate slices and their segmentations from 3 different patients are shown.
Discussion and Conclusions
In this work we have presented a framework for the segmentation of breast structures based on atlas. To the best of our knowledge this is the first proposal using atlas-based methodology for breast segmentation. Firstly we have constructed a probabilistic atlas by registering 8 patient data sets onto a single patient. Thereupon, we have integrated it into a Bayesian framework with MRF regularization for segmentation of breast MRI structures. Affine registration focused on VOI and B-Splines registration algorithms have been evaluated. The former has presented satisfactory results (general DSC average of 0.8) and acceptable execution time to be suitable for routine clinical use in the future.
For the pectoral muscle segmentation, which is the organ of main interest in this paper, the mean of DSC values is approximately 0.7 and good delineations have been obtained in intermediate slices. Even though the DSC value is not as good as for the other structures, this result is encouraging, especially considering that the pectoral muscle is a small structure compared with the others segmented and presents high inter-patient variability. We believe that the obtained segmentations are suitable as a reference tissue in pharmacoki-netic model calibration, where a specific percentage of voxel well labelled is needed.
Further research will be focused on solving the problems we found that affect the segmentation results. Firstly, we were aware of the low number of cases for the evaluation, strictly related with the difficulty of acquiring annotations for the 7 different classes in large volumes (108 slices per volume). However, evaluation will be extended to more cases by increasing the data set. Secondly, although bias field on images was corrected for, they still presented inhomogeneities. Thus, other correction methods will be studied. In addition, B-Splines registration has not provided good deformation alignment and has introduced additional registration errors. This could be explained by the fact that inter-patient variability is larger than the one which could be minimized by the implemented local registration. Other fast non-rigid registration algorithms, which incorporate user intervention such as control points or adding local deformation constraints, will be studied. Finally, signal intensity has not enough discriminative power to separate the pectoral muscle and glandular tissue classes. The use of other features, such as the ones obtained from Dynamic Contrast Enhanced breast MRI, and other voxel classification methods that use atlas information will also be considered.