Thrombin is the pivotal enzyme in the process of blood clotting or coagulation. It is the final proteinase of the proteolytic cascade, catalyzing the cleavage of two peptide bonds in its primary substrate fibrinogen, to generate fibrin, which subsequently polymerizes to form an insoluble clot. Thrombin has a multiplicity of additional functions, however, that contribute to both the amplification of blood clotting and its regulation. Thrombin promotes feedback activation of the coagulation cascade by activating the cofactors, factors V and VIII, as well as activating the proteinase zymogens, factors XI and XIII. Thrombin also stimulates the aggregation of blood platelets by activation of the thrombin receptor or PAR-1 (protease-activated receptor-1), one of four members of a sub-family of seven-transmembrane G-protein-coupled receptors, by a novel proteolytic "tethered ligand" mechanism (1). Thrombin also has anticoagulant properties and functions, because binding to the endothelial cell membrane protein thrombomodulin alters the specificity of thrombin, enabling it to activate another proteinase zymogen, protein C, which can, in turn, proteolytically inactivate factors V and VIII. Through activation of PAR-1, PAR-3, and PAR-4 thrombin can also mediate responses in a variety of cell types other than platelets, including endothelial cells, vascular smooth muscle cells and fibroblasts. Thus, despite its unique role in the generation of fibrin, thrombin can be viewed as a pleiotropic molecule, exhibiting both enzymatic and hormone-like properties.
Thrombin itself is generated by limited proteolysis of prothrombin, a 579 residue single-chain glycoprotein (Mr 71,600) expressed in the liver as the product of a 19 kb gene comprising 13 introns and 14 exons located on chromosome 11. Prothrombin undergoes important post-transitional modifications in addition to N-linked glycosylation at Asn 78, 100 and 373. The first 10 N-terminal glutamic acid residues, contained within a single 32-residue domain (known as the Gla domain), are modified to g-carboxyglutamic acid (Gla) by a vitamin K-dependent carboxylase, a modification common to all of the "vitamin K-dependent" coagulation factors, namely, factors VII, IX, X, protein C, and prothrombin. This modification is signaled for by residues -17 to -1 of the preprothrombin sequence, adjacent to the secretion signal peptide at -43 to -18. These Gla residues chelate Ca , leading to an ordering of the structure of the N-terminal structural domain, which is then able to bind to negatively-charged phospholipids. Prothrombin also contains two kringle domains that lack the lysine-binding properties of other kringle modules [see Plasminogen] and appear to have no discrete biological function, and the C-terminal serine proteinase domain. Structures for all the domains of prothrombin – specifically, fragment 1 (Gla-kringle), fragment 2 (kringle 2), and the serine proteinase domain – have been solved by X-ray crystallography and/or NMR. The structure of a-thrombin has been studied particularly extensively, in order to elucidate the molecular basis of its highly restricted substrate specificity.
The serine proteinase factor Xa in the prothrombinase complex [see Blood Clotting], activates prothrombin by cleavage at two positions (2). The first cleavage is at the Arg320-Ile321 peptide bond, generating "meizothrombin", a disulfide bond-linked two-chain proteinase that has a competent active-site, although lacking the specificity of mature thrombin. Subsequent cleavage at Arg271-Thr272 liberates the N-terminal fragment 1-2, which retains the phospholipid binding characteristics of prothrombin, plus fully mature a-thrombin. In the absence of cofactors, factor Xa cleaves the same two peptide bonds in the reverse order, first generating fragment 1-2 and the catalytically inactive prethrombin 2, which is subsequently activated to a-thrombin. The loss of the N-terminal domains on activation is unique among the blood coagulation proteinases and results in thrombin having full activity in the solution-phase, with no cofactor requirements.
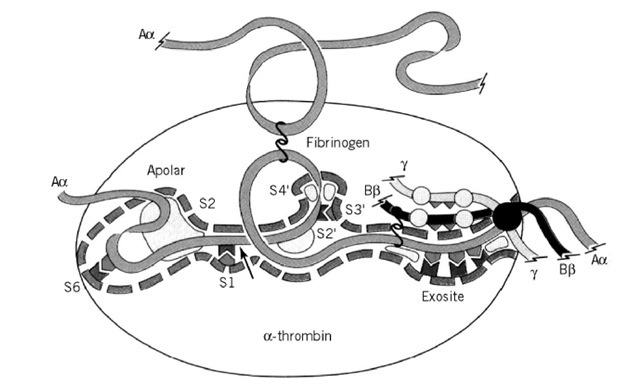
Thrombin consists of two disulfide-bridged polypeptide chains. The shorter A chain forms an integral part of the structure but not involved in catalysis. The B chain exhibits the characteristic fold of the trypsin-like serine proteinases, having two 6-stranded b-barrels with the equivalents of the catalytic residues Ser195, His57, and Asp102 (chymotrypsin numbering, corresponding to Ser525, His363, and Asp419 of thrombin, respectively) located at their interface. As with trypsin, substrate recognition involves a basic P1 residue, but thrombin has a much more extensive substrate recognition surface, as shown in Figure 1. Substrate recognition, and its very restricted specificity, is dictated largely by a number of unique insertion loops that border the active-site cleft, rendering it both deeper and narrower than that of chymotrypsin. These loops limit the access of many potential substrates and inhibitors to the active site, as well as imposing constraints on individual subsites. An example of this is the "60-loop" (a four-residue insertion at a position corresponding to Val60 of chymotrypsin), which restricts substrate P2 utilization to small hydrophobic residues, and forms part of the "apolar binding site" (3).
Substrate recognition by thrombin also involves the use of "exosites" to an unusually high degree.
Figure 1. The active-site of thrombin. Schematic diagram showing the major interactions of the fibrinogen Aa chain with thrombin and highlighting extensive area involved in substrate recognition.
These are regions of the active-site cleft far removed from the catalytic residues, the most significant of which is the "fibrinogen recognition exosite" (4). This is a cationic region to the "east" of the active-site (in the "standard orientation" shown in Figure 1). It is involved in catalytic interactions with fibrinogen and the thrombin receptor and non-catalytic interactions with thrombomodulin and the inhibitor hirudin (derived from the blood sucking leech Hirudo medicinalis). The latter is of particular interest as both the remarkable specificity and affinity (Kj ~10 fM) of this inhibitor are primarily dictated by this exosite interaction. The other major exosite is the heparin-binding site lying to the north-west of the active site (not shown in Figure 1), which is involved in the physiological inhibitory reaction with the serpin antithrombin bound to heparin or other sulfated glycosaminoglycans (5).
Many of the principles of substrate recognition elucidated in the extensive studies of thrombin will also apply to other serine proteinases with restricted substrate specificity; high resolution structures of them are also being solved.