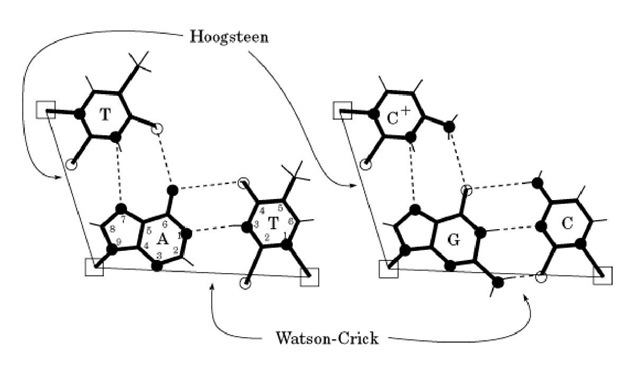
Nucleic acid bases can form a variety of base pairs. In all canonical duplex structures including B-DNA, A-DNA, and Z-DNA, the so-called Watson-Crick base pairs are found. The Watson-Crick base-pairings of guanine with cytosine and adenine with thymine are shown in Figure 1. The G to C and A to T complementarities are provided by specific hydrogen bond donors and acceptors between the bases. In a G:C base pair three hydrogen bonds are found between G-O6 and C-N4, G-N1 and C-N3, and G-N2 and C-O2 positions, and in an A:T base pair two hydrogen bonds are found between A-N6 and T-O4, and A-N1 and T-N3 positions. The two C1′ atoms within a G:C base pair and an A:T base pair are equidistant (~10.5 A). Thus the G:C and A:T base pairs in the Watson-Crick conformation are nearly iso-structural, from the point of the view of the sugar-phosphate backbone.
Figure 1. Schematic illustration of the A:T and G:C Watson-Crick and Hoogsteen base pairs. Hydrogen bonds are shown as dashed lines, and the distance between two C1′ atoms (shown as open squares) is drawn with a thin line. The numbering system of the purine and pyrimidine rings is shown.
In earlier crystallographic analyses of nucleic acid base pairs, it was found that an A:T base pair can also adopt a different conformation. Instead of using the N1 position of adenine to base pair with N3 of thymine, the N7 position of adenine was used. This type of A:T base pair was named Hoogsteen base pair (1). A striking difference between those two types of base pairs is that the C1′-C1′ distance in the Hoogsteen base pair of 8.65 A is significantly shorter than that of the Watson-Crick base pair of 10.5 A. Therefore the Hoogsteen base pair is not compatible structurally with the Watson-Crick base pair in B-DNA. The incorporation of a Hoogsteen base pair in B-DNA destabilizes the duplex structure. A similar G:C Hoogsteen base pair is not stable unless the C is protonated at the N3 position.
However, certain chemical modifications may enhance the stability of the Hoogsteen base pair. It was shown that the modified nucleoside 3-isodeoxyadenosine (iA) forms a stable base pair with thymine (T) using the Hoogsteen conformation, and the iA:T base pair is fully compatible with the normal Watson-Crick base pair as evident from the stable duplex of d(CG[iA]TCG) 2 shown by NMR analysis (2). Another modification which stabilizes the Hoogsteen base pair involves the use of 8-amino-adenine in which the amino group at the C8 position can form an additional hydrogen bond with the O2 of thymine (3). Those modified bases may find applications when the Hoogsteen base pairing is needed in nucleic acid structures.
The binding of protein or drug to DNA may induce alternative base pairs, including Hoogsteen base pair, to form. It has been shown that the quinoxaline bis-intercalator antibiotics triostine A and echinomycin bind to -XCGY- sequences with the drug bracketing the CG dinucleotide base pairs. In the structure of the triostine-CGTACG and triostine-GCGTACGCT complexes, the A:T and G:C+ base pairs adjacent to the quinoxaline rings adopt Hoogsteen base pairs (4). The molecular basis for the Hoogsteen base pair in the structure is to improve the stacking interactions between the quinoxaline ring and the adjacent base pairs. It is likely that such a conformational rearrangement from the Watson-Crick to Hoogsteen base pairs may occur in other biological systems.
An important role for the A:T and G:C+ Hoogsteen base pairs is found in the structure of a nucleic acid triple helix. The formation of a poly(A).poly(U).poly(U) triplex requires that the third strand of poly(U) is bound to the poly(A) strand of the poly(A).poly(U) duplex using Hoogsteen base pairing.
The T.A.T. and the C.G.C+ triple base pairs are shown in Figure 1. (Note that the triple C.G.C. base pair is not stable unless the second C is protonated.)
It should be pointed out that Hoogsteen base pairing is not restricted in DNA. In transfer RNA, the nucleic acid bases are often modified and involved in tertiary interactions. For example, in yeast tRNAPhe the nucleotide at position 58 is 1-methyl-adenosine, which is base paired with T54 (5). Because the N1 position of this adenine is methylated, it cannot participate in the normal Watson-Crick base pair. Instead m1 A58 and T54 form a reverse Hoogsteen base pair. It is likely that those noncanonical base pairs, including Hoogsteen and reverse Hoogsteen base pairs, will be found in higher-ordered RNA structures.