DNA replication in a chemical sense is the process by which an exact copy of a DNA molecule having a specific base sequence is synthesized. Exact copies of a linear DNA molecule can be replicated in vitro using purified DNA polymerases and proper primers. A recent advance in the long PCR method has made it possible to amplify DNA molecules as long as several tens of kilobases. In biology, however, DNA replication is defined as the duplication of the entire genome DNA in the cell. Basic mechanisms of replication of plasmids, bacteriophages, animal viruses, and bacterial chromosomes have been elucidated at the molecular level. On the other hand, knowledge of the replication of eukaryotic genomes is still limited, although increasing rapidly. A variety of structures and types of replication are known among plasmids, bacteriophages, plant and animal viruses. Mitochondria genomes are known to have a unique mode of replication. Here, replication of the genomes (conventionally called chromosomes) of prokaryotes (bacteria) and eukaryotes will be described, since the replication of genome DNA is a process essential for basic cellular functions, cell cycle, cell division, and cell differentiation.
1. Prokaryotic Genomes (Chromosomes)
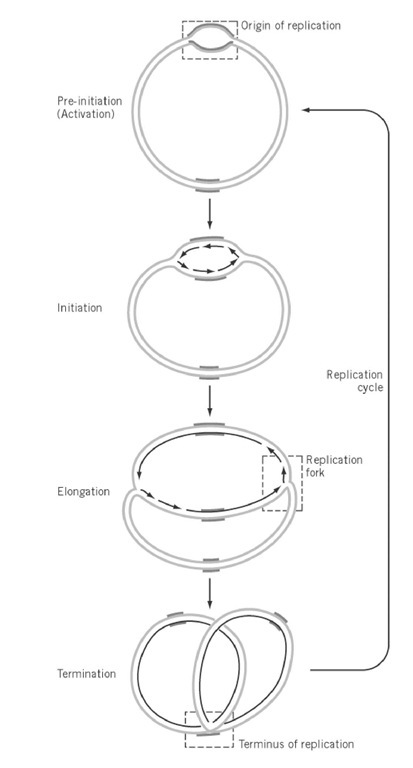
Bacterial genomes usually consist of circular DNA of one to eight megabases and comprise a single replication unit, a replicon. The general aspects of genome replication have been elucidated in Bacillus subtilis and Escherichia coli, representatives of Gram positive and Gram negative bacteria, respectively. In both genomes, replication is initiated from a genetically defined site, oriC (see Replication Origin), proceeds bidirectionally and symmetrically at about the same elongation rate, and terminates at the defined region, terC, of the genome (Fig. 1) (see Termination Of DNA Replication). Isolation of dna temperature-sensitive mutants has revealed that the three processes, initiation, elongation, and termination of replication, are regulated independently by multiple gene products. Genetic and biochemical studies subsequently revealed multiple protein complexes, the primosome and replisome, for the initiation and replication machinery, respectively (see DNA Replication Proteins). In contrast, termination is achieved by a single protein, Rtp.
Figure 1. Replication of circular prokaryotic chromosome. Replication of a circular chromosome is schematically shown. Replication is initiated by unwinding of a specific site of the chromosome, the origin of replication (oriC), followed by the synthesis of two leading strands and lagging strands in opposite directions. Elongation proceeds bidirectionally, with the same rate of synthesis at the two replication forks. Elongation terminates at a specific site, the terminus of replication.
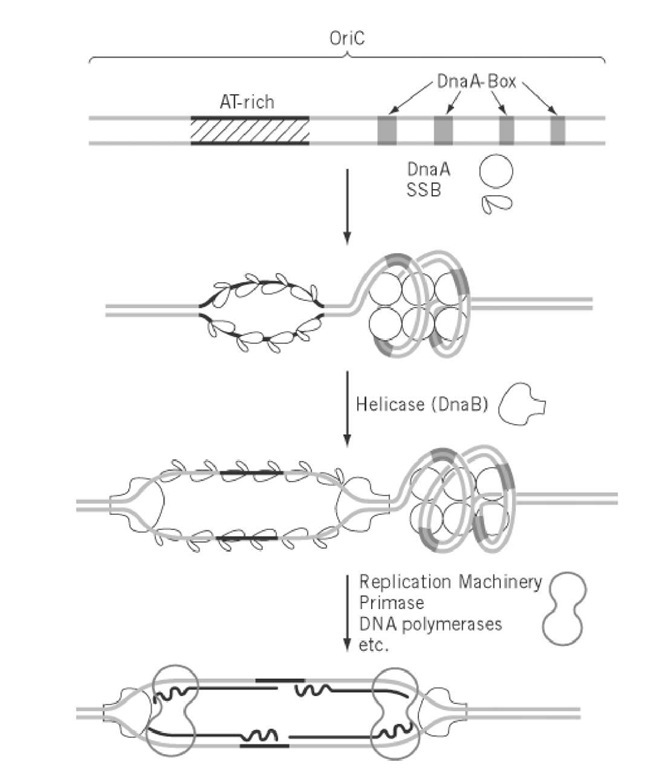
Regulatory DNA elements required for initiation, oriC, were identified by cloning of sequences that confer upon circular DNA the ability of autonomous replication as extra chromosomal elements in the cell (1). The oriC was first identified in E. coli, and it was then found that one of the E. coli Dna proteins, DnaA, functioned as an initiator of replication by binding to a sequence-specific element, the DnaA-box, to activate oriC and to unwind the AT-rich region within oriC (2). The combination of the DnaA-box and the DnaA protein functioning as cis and trans regulatory elements in the initiation of replication was subsequently found to exist commonly in many eubacteria (3). A replicon mechanism consisting of two regulatory elements, the replicator in cis and initiator in trans, that was proposed as early as 1963 (4) has been confirmed in prokaryotes (see Replicon).
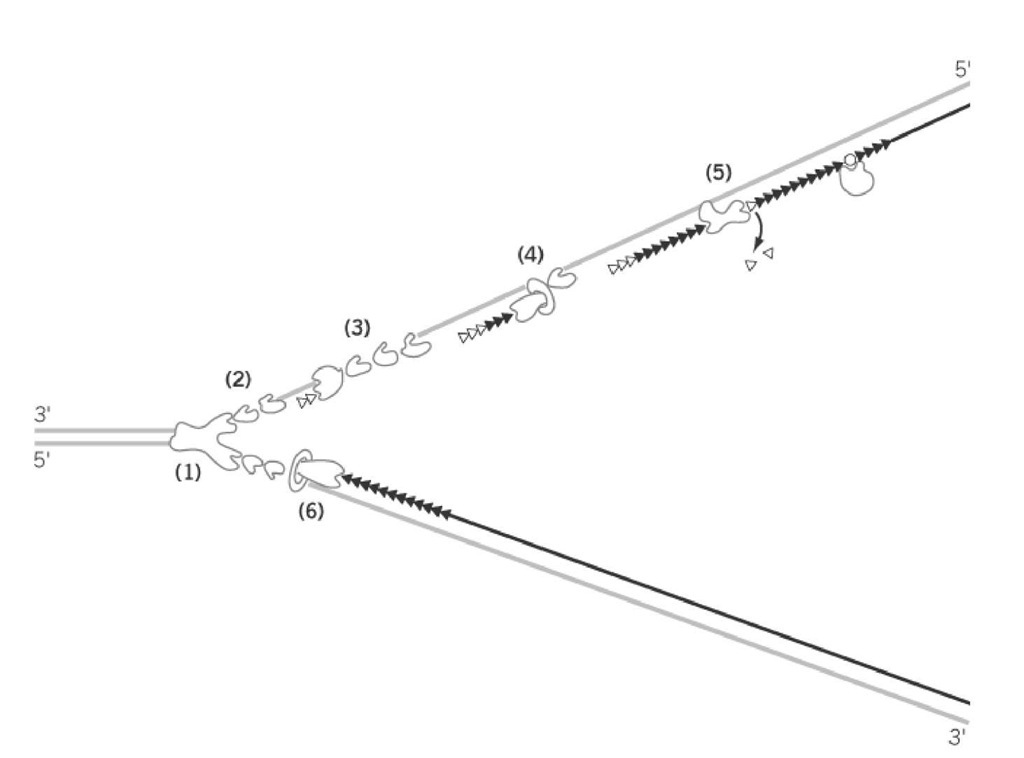
The elongation of replication is much more complicated than it was thought in the late 1950s when semi-conservative DNA replication was demonstrated and Kornberg discovered DNA polymerase I, which apparently copied double-stranded DNA in vitro. This complexity is due to the fact that the two strands of DNA are oriented in opposite directions in terms of the phosphodiester bonds and DNA polymerases can function in only one, the 5′ to 3′ direction. Extensive analysis of growing replication forks in E. coli led to the discovery of asymmetric discontinuous DNA replication in which synthesis of the 5′ to 3′ strand proceeds sequentially (the leading strand), whereas the opposite strand (lagging strand) is synthesized in short pieces, using an RNA primer, and subsequently joined together. The small RNA-DNA fragments of some 500 to 1000 bases synthesized during the discontinuous replication were named Okazaki Fragments after the researcher who discovered this mechanism (5). At least five enzymes, DNA helicase, primase, DNA polymerase I and III, and DNA Ligase, are involved in this process (Fig. 2) (see DNA Replication Proteins). These proteins are assumed to form a supramolecular complex, the replisome, and may be attached to the cell membrane. The initiation of replication is the synthesis of the first primer RNA-DNA molecule on both strands of DNA at the unwound region of oriC, which eventually become leading strands extended in both directions. The formation of the replisome at oriC requires the activation of oriC by DnaA and subsequent formation of the primosome complex, which includes the DNA helicase, DnaB in E. coli. The synthesis of lagging strands may be initiated after the formation of a considerable size of single-stranded region by elongation of the leading strand, but the detailed mechanism of synthesis of neither the first primer RNA nor the first lagging strands is known (Fig. 3).
Figure 2. Semiconservative and RNA-primed discontinuous replication. The mode of synthesis of leading strand and lagging strand at the replication fork is shown schematically. (1) Unwinding of double strand by helicase, (2) stabilizatio of single-stranded regions by single-strand DNA binding proteins (SSB) in prokaryotes (= p) or RPA in eukaryotes (= e) (3) primer RNA synthesis by primase (in p) or polymerase a-primase complex (in e), (4) lagging strand synthesis by DN polymerase III (in p) or polymerase d/e (in e), (5) degradation of primer rNa by polymerase I (in p) or ribonuclease H ( e), and (6) leading strand synthesis by polymerase III (in p) or polymerase d/e (in e).
Figure 3. Mechanism of initiation of replication of prokaryotic chromosomes. The OriC of E. coli and initiation from the oriC by the function of DnaA protein followed by DNA helicase and finally by assembly of the replication machinery is shown schematically. The DnaA box is a 9-mer sequence (consensus sequence is TTATCCACA) and DnaA protein is conserved in many eubacteria. The DnaB helicase is also conserved in at least E. coli and B. subtilis.
Elongation at the macroscopic scale proceeds bidirectionally along the circular genome at about the same rate, 50 kilobases/min, and completes at the fixed region, the terminus where two forks meet (s Termination Of DNA Replication). Two sets of three (a total of six) termination signals, oriented in opposite directions, are found in both E. coli (6) and B. subtilis (3) chromosomes at about 180o from oriC, to which a protein known as replication termination proteinein, Rtp, binds and inhibits the elongation in one direction. The distance between the nearest two signal sets is 270 kb in E. coli and 59 kb in B. subtilis. Although the mechanism of termination is basically the same in E. coli and B. subtilis, there is no homology in these terC sequences nor in the primary structures of Rtp. This is in sharp contrast to the extensive conservation of the DnaA protein and the DnaA-box sequence among eubacteria. The tertiary structure of B. subtilis Rtp protein reveals the mechanism of inhibition of DN replication at the molecular level (7). Termination of replication results in the two daughter chromosomes entwined about one another. Resolution of such a structure into two separate chromosomes can be achieved by the action of DNA gyrase (8) (see DNA Topology). In addition to the termination site, pausing sites where DNA replication slows down significantly are found near th termination sites of the E. coli chromosome, and their possible involvement in recombination has bee discussed (9).
2. Eukaryotic Chromosomes
Studies on the replication of a viral genome, SV40, have provided the basic knowledge about DNA replication in eukaryotic cells, because it replicates using host proteins, except for a single viral protein, T Antigen. T antigen was first identified as a protein responsible for tumor transformation ol mammalian cells by the tumor virus and hence was named as Tumor antigen. Subsequently, it was found that the T antigen is an initiator protein and a helicase that acts on the origin of the 3-kb circul SV40 genome (10). Success in reconstitution of an in vitro replication of the entire SV40 genome let to the identification and purification of enzymes and protein factors involved in the initiation and elongation of replication and separation of replicated molecules (see DNA Replication Proteins). Thi basic mechanism of synthesis of leading strand by DNA polymerase d/e and of lagging strand by the combination of the DNA polymerase a-primase complex and polymerase d/e has been elucidated (11 (Fig. 2). In addition, various factors, such as RPA, RFC, and proliferating cell nuclear antigen (PCNA), were identified to facilitate the efficient and progressive replication of the viral genome (12 A topoisomerase, topoisomerase II, was found essential for resolving the concatenated structure formed at the end of replication of the circular genome. Since no in vitro replication system is availa using cellular genome DNA as template, the molecular mechanism of replication established by the study of SV40 genome is still the sole model of eukaryotic DNA replication and serves as a unique system to identify factors involved in DNA replication and its regulation (see Replication Fork (Y-Fork Intermediate)) (13).
The mechanism of initiation of replication of the SV40 genome is similar to, and rather simpler than, that of bacterial genome, because T antigen acts as helicase as well as the initiator. However, it did n provide a model for the initiation of replication of cellular chromosomes composed of multiple replicons, which was first demonstrated by the autoradiography of replicating chromosomes in 1968 Huberman and Riggs (14). In general, the genomes of eukaryotic cells are estimated to contain aboui one origin every 10 to 330 kbp (15). Extensive searches for chromosomal replication origins through the cloning of autonomously replicating sequences (ARS) have mostly failed, except for yeast chromosomes of Saccharomyces cerevisiae and Schizosaccharomycespombe. A number of ARS hav been isolated from the 16 chromosomes of S. cerevisiae. In particular, 14 ARS in a 200-kbp portion chromosome III (16) and 10 ARS in the 300-kbp entire chromosome VI (17) were identified. All AR contain a common sequence of 11 bp ARS consensus sequence (ACS) that is essential for autonomo replication, plus additional nonspecific AT-rich sequences whose deletion causes a significant reduction in ARS activity. In some cases, a binding site for a transcriptional factor is located near 1 ACS and enhances its ARS activity. The ARS from S. cerevisiae is unique, and no homologous sequence of the approximately 100-bp ACS was found in other eukaryotes. ARS cloned from the oth yeast S. pombe require a region of about 1 kbp containing several AT-rich clusters, with no single specific sequences like ACS essential for ARS activity. The two-dimensional gel electrophoresis of chromosomal fragments developed by Brewer and Fangman (18) has successfully detected the eye-form intermediates of replicating intermediates clearly separated from Y-form intermediates (see Replication Fork (Y-Fork Intermediate)). Using this method, some ARS are found to function as efficient origins of chromosomal replication in S. cerevisiae. However, some of the active ARS in plasmids were found to be very inefficient or silent on the chromosome, suggesting a role of chroma structure in origin activity. Not all origins are fired at the same time in S phase, but they are initiatec sequentially in a fixed order (19). A regulatory factor determining the efficiency and timing of initiation of late replicating origins have been identified (24).
Origins from genomes of higher organisms are ambiguous. Attempts to isolate ARS from mammalia chromosomes are difficult to reproduce and therefore controversial. The best-studied origin, near the dihydrofolate reductase (DHFR) gene in the Chinese hamster genome, showed variation in size from 0.5 to 55 kbp, depending on the methods used to detect origin activity (20). Systematic studies on lar chromosomal segments have revealed that most fragments longer than 10 kbp can provide some AR activity in mammalian cells, suggesting that DNA length is more critical than DNA sequence. In man cases, initiation occurs randomly within several kilobases, which is called the initiation zone rather than the origin. The concept of a replicator that is proposed in the original replicon hypothesis should be reexamined in these complex origins, in terms of the recognition site of initiators and the actual si of initiation of replication (see Replicon).
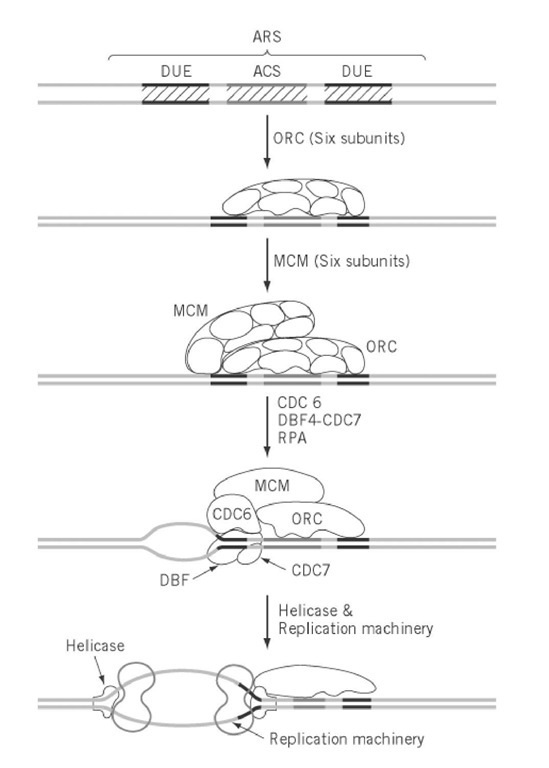
As for trans factors for initiation, no single initiator protein like bacterial DnaA or SV40 T antigen h been found in eukaryotic cells. Instead, a complex consisting of six proteins was found to recognize yeast ARS through binding to the ACS and named accordingly as the origin recognition complex (ORC) (21). The ORC was subsequently found to be well conserved from yeast to humans. Howevei the mechanism of how ORC recognizes seemingly nonspecific origin sequences on chromosomes other than S. cerevisiae is not clear. An in vitro DNA synthesis system using a Xenopus egg extract and sperm chromosomes provided a unique system for studying the biochemistry of initiation of chromosomal replication, and it led to the discovery of the licensing factor that permits replication o: multiple replicons of the chromosome once in the cell cycle (22, 23). Genes homologous to the Xenopus licensing factor were identified among MCM genes of S. cerevisiae, and a MCM complex composed of six proteins that binds to ARS through interaction with ORC was subsequently discovered. Genetic studies with S. cerevisiae show that the formation of the MCM-ORC-ARS complex is not sufficient to activate the origin of the chromosome, and additional factors, protein kinases that interact with MCM or ORC directly or indirectly, were being discovered. Extensive studies on the regulatory network of the replication complex are expected to elucidate the molecular mechanism of signal transduction from the cell cycle engine, cdk/cyclin kinase, to the chromosomal origin of replication. The MCM complex, as well as the factors interacting with MCM and ORC, are conserved widely in eukaryotes, suggesting that the molecular mechanism elucidated in the yeast and frog provides principles that guide the studies on more complicated chromosomes including human chromosome (Fig. 4).
Figure 4. Mechanism of initiation of replication of a Saccharomyces cerevisiae chromosome. Minimal structure of the ARS contains a 11-bp sequence-specific region (ACS) that is essential for the origin, and surrounding stimulative regio that are rich in AT and called DNA unwinding elements (DUE). The ORC binds to ACS throughout cell cycle. MCM (licensing factor) complex binds to ORC at the G2-M phase of the cell cycle, and the double complex may be activated by several protein kinases, including CDC6 and DBF4/CDC7, to produce a single-stranded region that serves as the ent site for helicase and eventually for the replication machinery. The mechanism is hypothetical, as the last two steps have not been proven experimentally.
Termination of replication of adjacent replicons occurs when two forks moving in opposite direction meet. Although specific termination sites between the two adjacent replicons have not been identifie the structure of replicated replicons at the termination sites must be similar to the ends of the circulai chromosomes of bacterial and viral genomes and require topoisomerases to resolve the structure to separate two sister strands. Termination of the ends of linear chromosomes requires a more specific structure, the telomere, to prevent shortening of the chromosome due to the discontinuous replicatioi mechanism.