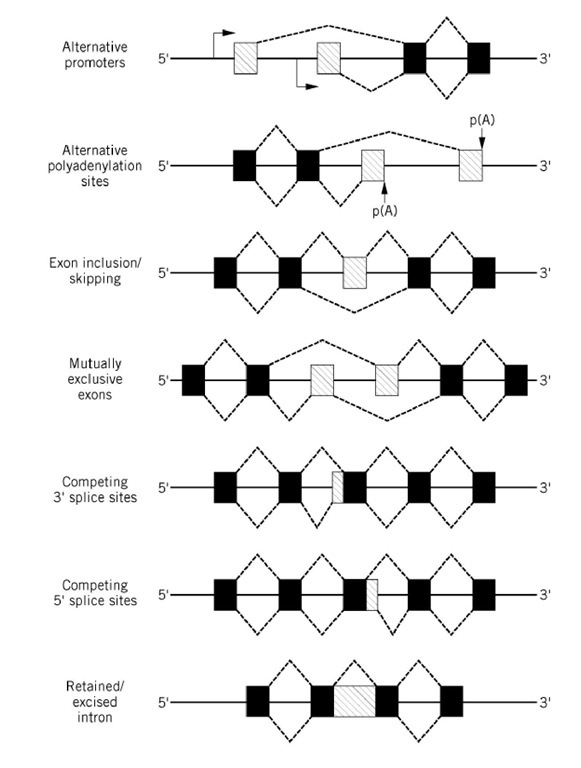
Most eukaryotic genes that code for proteins contain noncoding sequences (introns) that are interspersed among the coding regions (exons). Transcription of these genes generates so-called pre-mRNA molecules, which are converted to mature mRNAs by a process termed RNA splicing. During this process, the introns are precisely excised and the exons are ligated together (see RNA Splicing). The majority of nuclear pre-mRNAs are spliced constitutively; that is, only one mature mRNA species is generated from a single pre-mRNA in all tissues. In some cases however, alternative 5′ and/or 3′ splice sites are used during splicing, resulting in the production of more than one mRNA species from a single pre-mRNA. Alternative splicing has been documented for many eukaryotic genes, and a variety of alternative splicing patterns have been observed, as depicted schematically in Figure 1. The utilization of alternative 5′ and/or 3′ splice sites (also referred to as donor and acceptor sites, respectively) can result in structurally distinct mRNAs by either excluding potential exon sequences or incorporating otherwise noncoding intron sequences. For some pre-mRNAs, alternative splicing is a nonregulated event such that two or more alternatively spliced mRNAs are produced at a given ratio to one another in all cell types. For others, the choice of alternative splice sites is regulated in a tissue-specific or developmental manner. This type of regulation is mediated by trans-acting factors that are differentially expressed in a particular tissue or at a specific time during development (see text below). These trans-acting factors may be positive or negative regulators that activate or repress the use of an alternative splice site either directly or indirectly, for example by modulating the affinity of general splicing factors.
Figure 1. Patterns of alternative splicing. Constitutively spliced exons are shown as black boxes, alternatively spliced exons are shown as shaded boxes, and introns are shown as a solid lines between the exons. Transcription start sites are indicated by an arrow, and polyadenylation sites are indicated by p(A). Splicing events are depicted by a dashed line.
Alternative splicing can lead to both quantitative and qualitative changes in gene expression. Quantitative changes can arise if the alternatively spliced mRNA contains a prematurely truncated open reading frame (ie, due to the presence of a stop codon) or exhibits an altered stability or translation efficiency. In many cases, alternative splicing leads to the production of so-called protein isoforms that are structurally identical in all but a specific region or domain (1). Such structural variants of a given protein often exhibit significant functional differences. Thus, through the generation of multiple protein isoforms, alternative splicing can enhance the phenotypic variability of a single gene.
A central question in both constitutive and alternative splicing that has yet to be clearly resolved is how the correct pairs of 5′ and 3′ splice sites are selected for cleavage and subsequent ligation (see also Splice Sites). Pre-mRNAs contain multiple authentic 5′ and 3′ splice sites, which must be properly paired in order to prevent the random skipping of one or more exon. Furthermore, these sites must be distinguished from other nonauthentic sites that are also repeatedly present in most pre-mRNAs. In the case of alternative splicing, several different factors appear to be responsible for the preferential selection of one splice site over another. Firstly, features of the pre-mRNA itself (so-called cis-acting elements) can influence splice-site utilization. For example, the conserved sequences found at the 5′ and 3′ splice sites and the branch site contribute to splice site selection (see Splice Sites). In recent years it has become clear that the relative strengths of competing splice sites is often a deciding factor in determining which site is used preferentially. The strength of a particular 5′ or 3′ splice site is generally a measure of how efficiently it binds spliceosomal components, such as the U1 and U2 snRNPs or the splicing factor U2AF, which play important roles during the early stages of spliceosome assembly. This in turn is often, but not always, a function of how closely its sequences match the 5′ and 3′ splice site consensus sequences, or a function of the length and uridine content of the polypyrimidine tract, which determines its affinity for U2AF. The selection of weak 5′ and 3′ splice sites, on the other hand, can be enhanced by splicing factors that promote U1 or U2 snRNP binding (eg, SR proteins; see text below). Some 5′ splice sites also activate usage of an upstream 3′ splice site (eg, in the preprotachykinin pre-mRNA); and, vice versa, some 3′ splice sites can promote the use of a downstream 5′ splice site (eg, in the B-tropomyosin pre-mRNA) (2, 3). In the former case, factors bound at the 5′ splice site (the U1 snRNP) enhance the interaction of U2AF with the upstream 3′ splice site via a network of molecular interactions across the exon (4, 5)
In some cases, regions of the pre-mRNA other than the 5′ and 3′ splice sites also contribute to alternative splice-site selection. Purine-rich sequences (so-called splicing enhancers), which are typically located in exons and often bind SR proteins, are known to enhance the use of adjacent 5′ or 3′ splice sites by stabilizing the interactions of spliceosomal components with them (reviewed in Ref. 6). In some instances, intron sequences or sequences in the 3′-untranslated region that regulate polyadenylation have also been shown to influence alternative splicing. Finally, pre-mRNA secondary structure (eg, stem-loop structures) can affect splice-site selection, for example by blocking the interaction of spliceosomal components with a particular splice site (7, 8).
Splice-site selection during alternative splicing is also regulated by /raws-acting factors. Many of the currently identified trans-acting factors also function in constitutive pre-mRNA splicing. In this case, variations in their concentration, or the concentration of factors that compete with them, are thought to lead to tissue-specific modulation of splice-site usage. Foremost in this catagory of factors are members of the evolutionarily conserved SR protein family, which are characterized by an amino-terminal RNA-binding domain and a C-terminal domain rich in arginine-serine (RS) dipeptides (reviewed in Ref. 9). SR proteins (eg, SF2/ASF and SC35) play essential roles in constitutive nuclear pre-mRNA splicing, particularly at the earliest stages of spliceosome assembly (see Spliceosome). Moreover, in pre-mRNAs that contain multiple 5′ or 3′ splice sites (see Fig. 1), high concentrations of SR proteins generally enhance the use of the more proximal (downstream) 5′ splice site (so-called switching activity) or more proximal 3′ splice site (9, 10). However, apparently due to differences in the affinities of individual SR proteins for different pre-mRNAs, the effect of a particular SR protein on 5′ splice site selection can vary from one pre-mRNA to the next (11, 12). SR proteins appear to act by facilitating the interaction of the U1 snRNP with the 5′ splice site, an initial step for 5′ splice-site recognition, or by promoting the association of U2AF with the 3′ splice site (see Spliceosome). High concentrations of SR proteins can also inhibit exon skipping (Fig. 1), which likewise involves promoting the use of a proximal 5′ splice site. Interestingly, the activity of SR proteins such as SF2/ASF or SC35 in 5′ splice-site selection (but not in constitutive splicing) can be antagonized by the hnRNP proteins A/B (13, 14). High concentrations of these proteins generally favor the use of more distal 5 splice sites, and the ratio of 5 splice site usage is determined by the relative amounts of hnRNP A/B and SR protein. Thus, the amount of some alternatively spliced pre-mRNAs can be modulated by varying the cellular concentrations of these proteins (15). SR proteins are also involved in activating weak 5 and 3 splice sites that are located adjacent to purine-rich splicing enhancers (reviewed in Refs. 9 and 16). The binding of specific SR proteins to these exon enhancer sequences promotes the interaction of U2AF, and thus the U2 snRNP, with the 3′ splice site or the U1 snRNP with the 5 splice site.
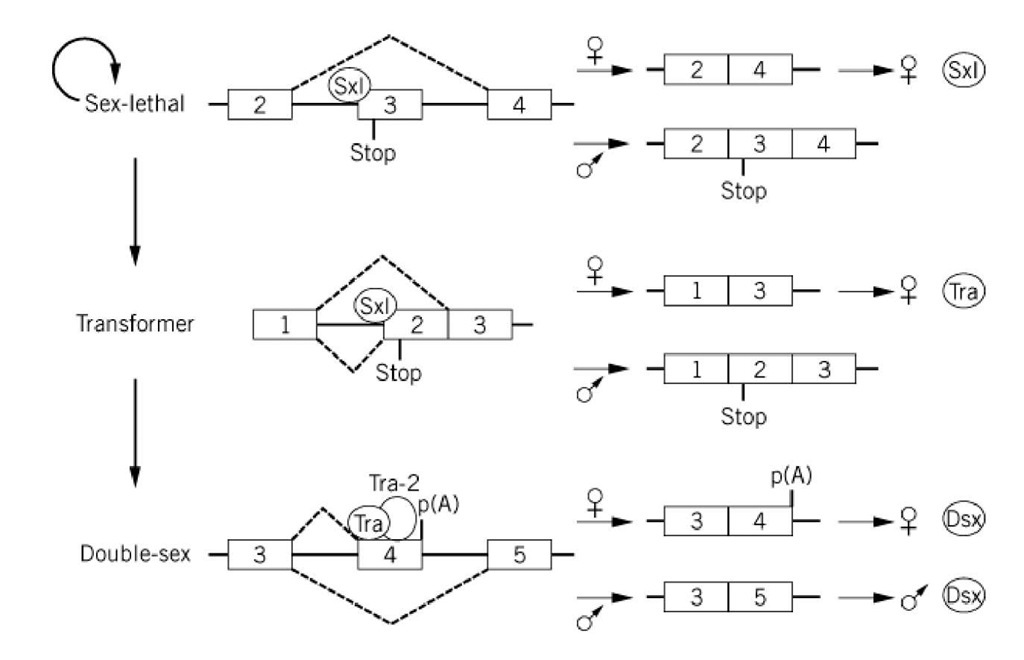
Trans-acting alternative splicing factors that are expressed in a tissue-, sex- or developmental-specific manner have also been identified. In mammals, concrete, well-understood examples of cell-specific regulators of alternative splicing are currently limited. The best-characterized factors are those responsible for alternative splicing events in the fruit fly Drosophila melanogaster. For example, sex determination in fruit flies involves a cascade of alternative splicing events that are regulated by sex-specific proteins (17) (see Fig. 2). In males, splicing of the Sex-lethal (Sxl), transformer (tra) and doublesex (dsx) pre-mRNAs are nonregulated events that appear to require only the general splicing machinery (ie, they represent the so-called default splicing patterns). In females, alternative splice sites are activated by female-specific factors, either directly or indirectly, through the inactivation of the male-specific site. The first of these factors known to act in this cascade is the female-specific Sxl protein, which regulates its own synthesis by an alternative splicing event. In females, Sxl is thought to block (by a currently unknown mechanism) use of the 3′ splice site of the third exon of the Sxl pre-mRNA, which leads to exon 3 skipping (18, 19). Because exon 3 contains a stop codon, functional Sxl protein is produced only from the female-specific Sxl mRNA (Fig. 2). In the next step of this cascade, the Sxl protein inhibits the use of the upstream 3′ splice site of exon 2 of the tra pre-mRNA, which in turn activates splicing at a weaker downstream 3′ splice site. Specifically, Sxl has been shown to bind to the stronger polypyrimidine tract of the upstream 3′ splice site and thereby to inhibit the binding of U2AF (20). This results in U2AF binding to the downstream site, for which it has a lower affinity. The resulting exclusion of exon 2, which also contains a stop codon, leads to the production of functional tra protein in females. In the last step, the tra protein, in conjunction with tra-2, directly activates use of the weak 3 splice site of exon 4 of the doublesex pre-mRNA. Tra and Tra-2, both of which contain RS domains, interact with a purine-rich splicing enhancer present in exon 4 and recruit SR proteins, as well as U2AF, to the upstream 3′ splice site (reviewed in Ref. 6). As a result, exon 4 is included in female-specific dsx mRNA, and polyadenylation occurs at the end of this exon. The resulting female dsx protein represses male differentiation, whereas the male protein represses female differentiation. Although in this particular case much has been learned about the molecular mechanisms responsible for splice-site selection, in most cases a clear understanding of the complex processes of alternative splicing awaits further investigation.
Figure 2. Cascade of alternative splicing events in the sex determination pathway of Drosophila. For simplicity, only those exons and introns involved in alternative splicing events are shown. In female flies, the sex lethal protein regulates its own synthesis and that of the tra protein by blocking the use of the 3′ splice site of the third and second exons of the sex-lethal and tra pre-mRNAs, respectively. Both of these exons contain a stop codon, as indicated. The tra protein, together with tra-2, interacts with a splicing enhancer in the fourth exon of the double-sex pre-mRNA and activates use of the upstream 3′ splice site. Exons are depicted as numbered boxes, and introns are depicted as solid lines. Dashed lines above the introns represent splicing events in females, and those below indicate splicing events in males. The sex-lethal (sxl), transformer (tra), and tra-2 proteins are indicated by circles. Polyadenylation is indicated by p(A).