1. Introduction
In 1989, while a Ph.D. student at the University of Oxford, Paul Dear invented a new method for genome mapping, named “Happy mapping”, which reflected the use of polymerase on minute amounts of DNA in the procedure. This method is basically an in vitro adaptation of the radiation hybrid (RH) method (Cox etal., 1990) in which random subsets of a genome of interest are integrated into the nucleus of a carrier cell to produce a panel of 80 or more independent hybrid cell lines.
This method offers several advantages over the RH method. First, it is possible to apply the Happy mapping method to any genome, including plant genomes, as no cell fusion is involved; second, a Happy panel can be produced in a few weeks as compared to several months for an RH panel; third, a Happy panel contains only the DNA of interest, which makes the analysis of markers easier as there is no possibility of interference with the genomic DNA of the carrier cell. Finally, the final computation of the vectors is simplified, and the resulting map is more robust as a result of the higher and more uniform retention value in each microtiter well of the Happy panel as opposed to the hybrid cell panel.
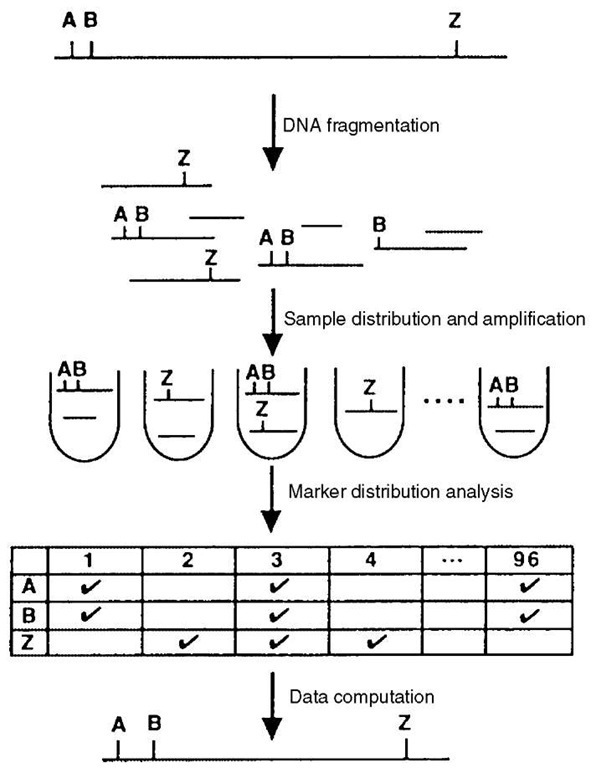
In the Happy method, minute amounts of genomic DNA, corresponding to less than a haploid equivalent of the genome of interest, are placed in the wells of a microtiter plate (Figure 1). Given the mass of a diploid mammalian genome (~5pg per nucleus), this corresponds to an average of 2pg of DNA per well. As a consequence of the limited amount of DNA in each well, only a subfraction of the whole genome is present and, as in the case of radiation hybrid cells, markers located close to each other on the genome tend to be found in the same wells of the microtiter plate. As for RH panels, the ability of a Happy panel to link two markers depends upon the size of the DNA fragments in the wells and, as a rule of thumb, the distance between two markers cannot exceed one-third to one-half of the mean size of the DNA fragments for them to be linked. Owing to the difficulty involved in manipulating very large DNA molecules, the Happy method tends to construct dense maps with at least one marker every megabase. To overcome this limitation, the construction of a Happy panel usually starts by incorporating entire cells or nuclei into agarose beads. The DNA is then gently extracted directly from these beads and subjected to pulsed-field gel electrophoresis. Following migration, bands of the appropriate size – fragments bigger than a few Mb are difficult to obtain -are excised and placed in the wells of a microtiter plate. A series of preliminary experiments are usually necessary to adjust the quantity of DNA placed in each well (between 0.5 and 0.9 haploid genome equivalents). Up to this point, a panel can be readily constructed for any cell type and only requires a bit of practice.
Figure 1 Schematic representation of the Happy mapping method
The next stage in map construction is to analyze the distribution of the markers within the panel. This is generally done by PCR as for RH mapping. The immediate advantage of a “Happy panel” as compared to an RH panel is the absence of foreign DNA that could interfere with the DNA of interest during amplification. However, the limited amount of DNA raises a specific problem as the detection of one marker requires two PCR with two pairs of nested primers and a multiplex approach would allow the analysis of a small number of markers only. To overcome this limitation, a whole-genome PCR is required. This poses technical problems that have not yet been satisfactorily resolved. An efficient PCR should satisfy quantitative and qualitative goals. DNA must be amplified 107- to 108-fold to get enough material for the further mapping of 103 or more markers. Even more importantly, the amplification has to be unbiased, that is, the composition of the DNA after amplification must correspond to its initial composition, such that all the markers present in a well before amplification are present afterward. During the last 10 years, several techniques using different cocktails of oligonucleotide primers and enzymes have been described, but none has met these requirements (Telenius et al., 1992; Zhang et al., 1992). In these studies, randomness of the product was obtained when a minimum of 30 to 50 DNA molecules was used, but not with just one molecule as in the case of a “Happy panel”. Furthermore, the amount of DNA obtained in these studies was limited, prohibiting direct use of the DNA in Happy mapping. Through reamplification of the PCR products, sufficient material could be obtained, but as a sizable fraction (between 30 and 40% depending on the Happy panel) of the markers could not be mapped accurately, PCR-induced representational bias was suspected (De Ponbriand et al., 2002).
As a proof of concept of their method, Dear et al. (1998) mapped 1001 markers from human chromosome 14. To overcome the PCR bottleneck, they performed inter-Alu PCR with one degenerate primer specific to the repetitive Alu sequence. Although this provided excellent proof of principle, as the resulting map was subsequently shown to match the corresponding human sequence, this sort of map is of little value to gene hunters as these marker sequences are usually nonpolymorphic and not readily usable for synteny comparisons.
It is striking to note that apart from Paul Dear’s group and ourselves, no one has ever published a map based on the “Happy technique” despite the potential advantages of this method. Owing to the absence of methods for obtaining sufficient quantities of unbiased amplified haploid DNA, the method has not been used to map any large genomes. Instead, Paul Dear and colleagues have produced genome maps of unicellular eukaryotes of less than 20 Mb (Abrahamsen et al., 2004) and of specific chromosomes such as Dictyostelium discoideum chromosome 6 (Konfortov et al., 2000). This involved a rather cumbersome two-step amplification approach. The first amplification with a limited yield was done according to Zhang et al. (1992) with a random 15-mer primer. In the second amplification step, aliquots of the first amplification product were subjected to multiplex PCR with between 20 and 200 pairs of primers corresponding to the PAC end sequences and additional markers (Piper et al., 1998). Finally, using an aliquot of this second amplification, marker-specific PCR was carried out to analyze the distribution of each marker within the Happy panel.
2. Future trends
Efforts to develop a PCR method that results in adequate yields and that can randomly amplify a minute amount of genomic DNA have been moderately successful.
However, the limited DNA yield obtained with the technique described by Zhang et al. (1992) has led Paul Dear and collaborators to develop a strategy on the basis of a two-step amplification combined with a multiplex PCR approach to map only a few hundred markers (Piper et al., 1998; Konfortov et al., 2000). Nevertheless, the potential of the approach and the robustness of the “Happy map” have been well established. Other recent developments, such as those based on DNA microarray technology or the identification of new enzyme activities, should rekindle interest in the Happy method and lead to the proposal of novel applications.
The development of dedicated microarrays and the possibility of reducing the complexity of large genomes by only amplifying small restriction fragments (Kennedy et al., 2003) should lead to new opportunities. It may, for example, be possible to spot PCR fragments corresponding to markers of interest (i.e., corresponding to short restriction fragment produced by a six-nucleotide cutter enzyme) onto functionalized glass and to hybridize them to fluorescently labeled DNA extracted from the different samples of a Happy panel. This approach would detect markers present in each sample instead of asking in which samples each marker is present. If properly developed, it should be possible to use this strategy to derive dense maps of mammalian genomes for which we will shortly have access to sequence information generated by low-pass shotgun sequencing. Other amplification strategies will certainly be developed using the QBeta polymerase sold by several companies and which has been shown to support high yields of random amplification. This enzyme could be used either alone or in combination with the amplification method described by Zhang etal. (1992). The attractive possibility of dissecting identified DNA regions or chromosomal bands under a microscope (metaphase spreads) should make it possible to prepare localized markers. It should then be easy to map these limited number of markers on a Happy panel that could be constructed using the currently available method in a few weeks rather than in a few months for an RH panel. These are several possibilities that remain to be investigated now that it has been well established that the Happy mapping method can produce robust maps and could thus be a powerful alternative to the RH method.