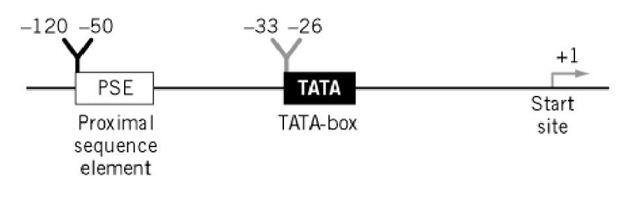
In higher eukaryotes, comparison of DNA sequences of a variety of RNA polymerase-II promoters revealed an A + T-rich consensus sequence, closely resembling the -10 Pribnow Box of bacterial promoters and centered approximately 30 base pairs (bp) upstream of the start site of transcription. This 8-bp element is called the TATA box because the maximum homology covers the first four bases, TATA (Fig. 1). It is also found in lower eukaryotes (such as Saccharomyces cerevisiae), although at variable distances from the start site, ranging from positions -30 to -120. In vivo as well as in vitro studies revealed the importance of this sequence for the initiation of accurate transcription.
Indeed, using in vitro transcription assays, it was shown that deletions encompassing part, or all, of the TATA sequence are detrimental for transcription (1, 2). Even a single base change within the TATA box drastically decreases transcription, indicating that the rate of transcription could be a function of the nature of the TATA-box sequence (3). The TATA sequence is involved in the selection of the transcription initiation site; accordingly, deletions of the original start site that leave the TATA box unchanged do not prevent transcription initiation, which still occurs approximately 30 bp downstream of the TATA element.
Figure 1. Organization of eukaryotic class II promoters.
Transcription requires correct positioning of RNA polymerase II on the promoter, in addition to other components called basal (or general) transcription factors. These factors participate in the formation of the preinitiation complex, which is nucleated by the binding of one of them, TFIID, on the TATA box. Targeting of the TATA box by TATA binding protein (TBP), a subunit of TFIID, allows the arrival of the other factors, either in a chronological (sequential) order or in a single step as a large complex, the RNA polymerase II holoenzyme.
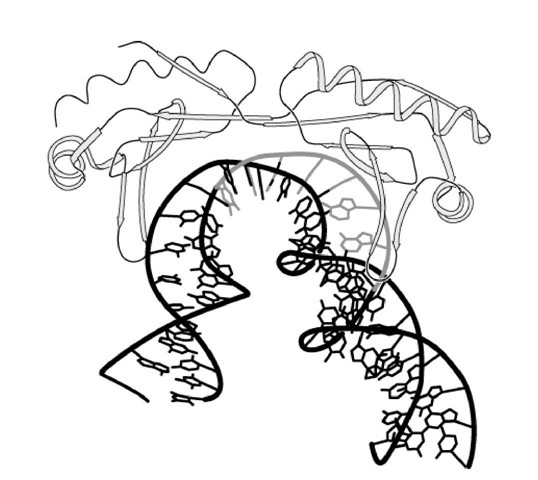
X-ray crystallography studies show that the carboxy-terminal domain of TBP consists of two direct repeats of about 80 amino acid residues and resembles a saddle that sits astride the DNA (4, 5), contacting its minor groove (Fig. 2). Insertion of two phenylalanine residues of TBP between the two last base pairs of the TATA element induces a bend of about 80° in the DNA structure, which widens the minor groove but does not impede the base pairing (6). Bending the DNA around the TATA box allows contacts between regulatory factors bound to their specific sequence, on one side, and the basal transcription machinery on the other side. It could also prevent condensation of DNA in chromatin structure and repression of transcription. The importance of the DNA kinking varies, depending upon the nature of the TATA box and the surrounding sequences.
Figure 2. View of the TBP-TATA complex. The TATA sequence is shown in dark stippling, whereas the surrounding DNA sequences are in bold lines. TBP, which is displayed in light stippling in a ribbon representation, induces the kinking of the DNA (9).
The affinity of TBP for the TATA element probably determines the strength of the corresponding promoter—a strong promoter allowing a high rate of transcription. It is still unclear, however, whether TBP recognizes the primary TATA-box sequence or simply its three-dimensional structure. Such a structural mode of recognition has already been suggested for class I promoters, which lack a canonical TATA box. Even though DNA bending at promoters is induced by the TATA-TBP interaction, recent data demonstrate that TBP is able to bind in a lock-and-key fashion to a similar bent structure provided by DNA damage, such as cisplatin adducts or ultraviolet photoproducts (7). The requirement of TBP for transcription from a TATA-containing promoter has been challenged by the discovery that a complex of TBP-associated factors (TAFs) without TBP (called TFTC for TBP-free TAFn-containing complex) could substitute for TFIID (8). TFTC does not bind the TATA-box itself, however, raising the possibility that another component does so during TFTC-mediated transcription.