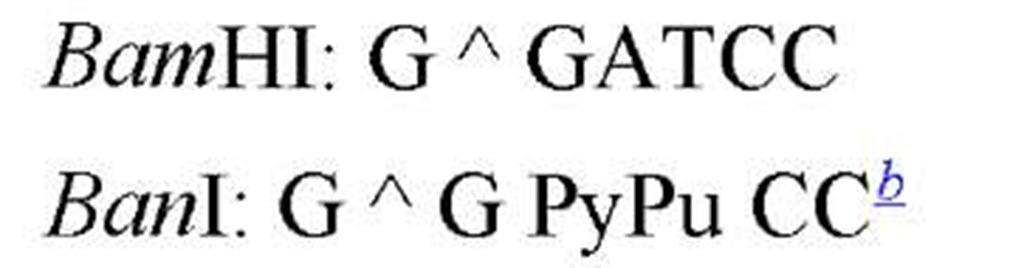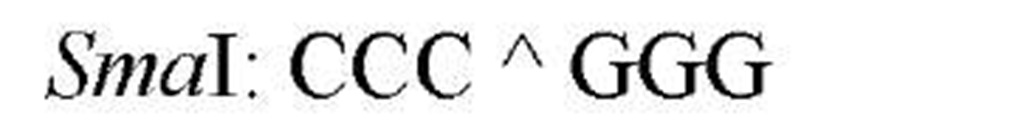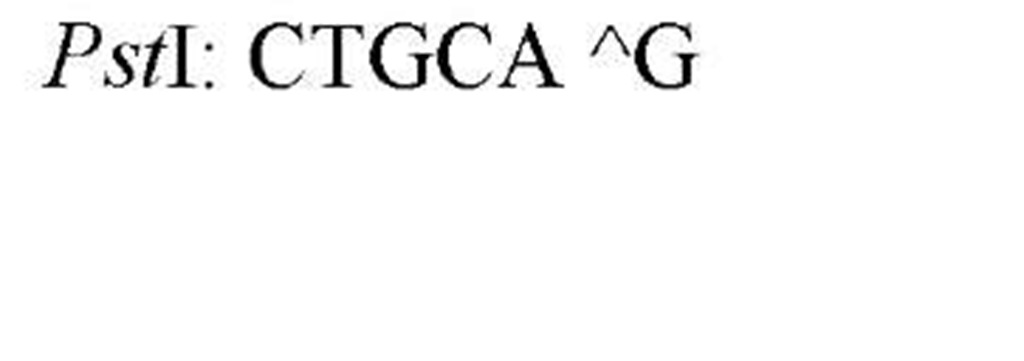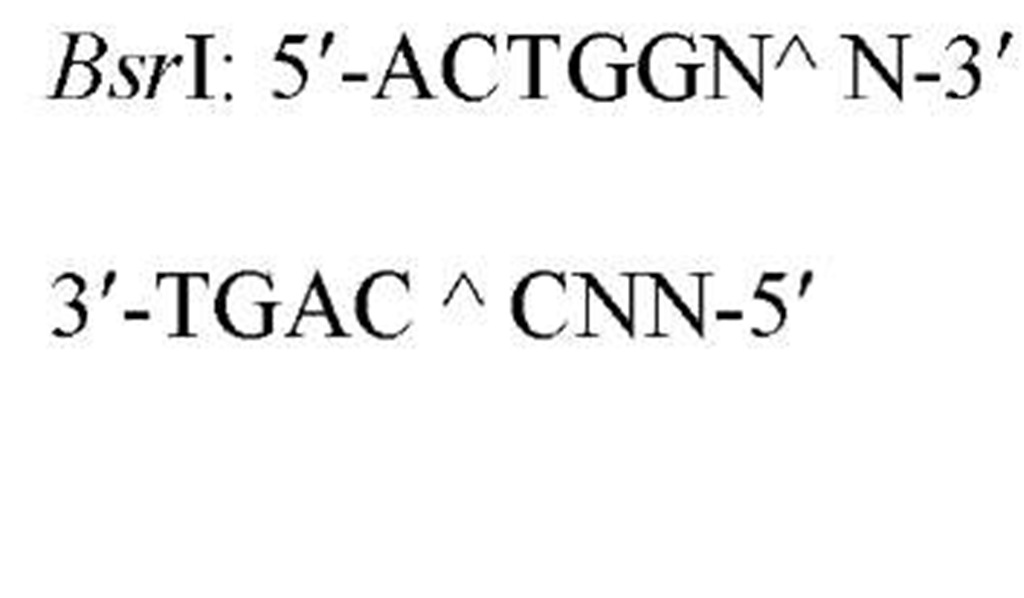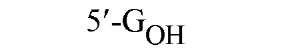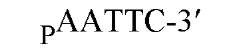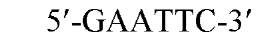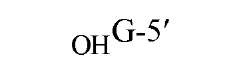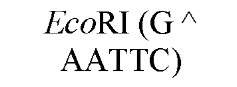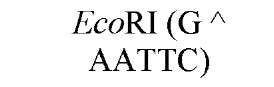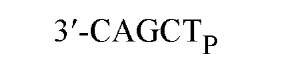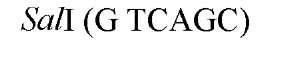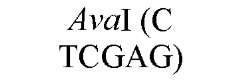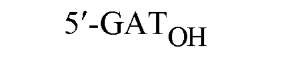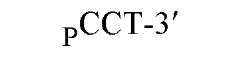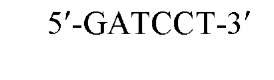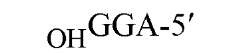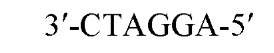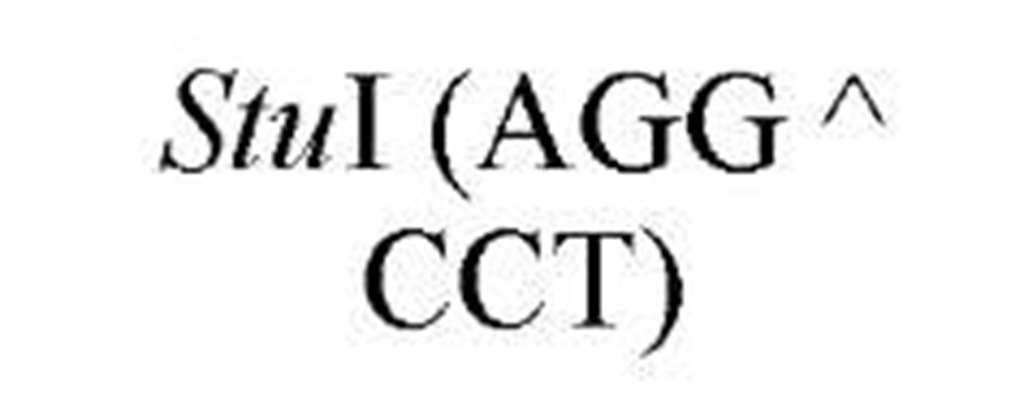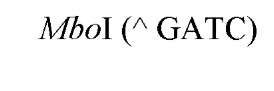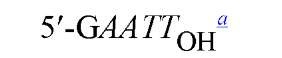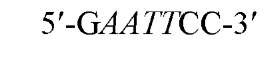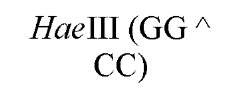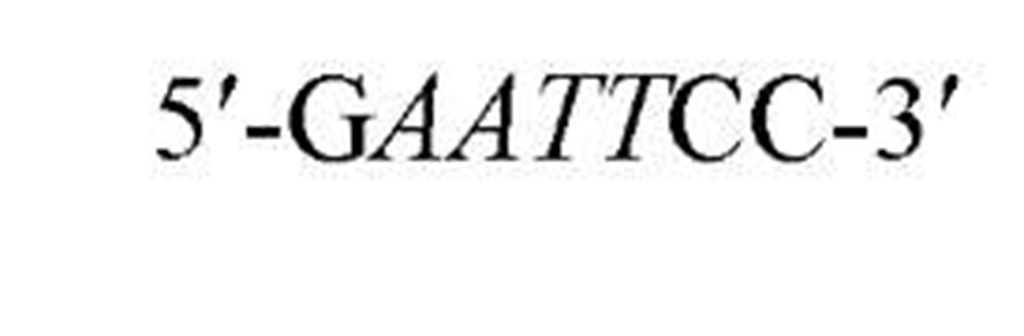Restriction Enzymes are bacterial endonucleases that recognize defined sequences of double-stranded DNA and catalyze phosphodiester bond hydrolysis of both strands at positions within or adjacent to the recognition sequence (see Restriction Enzymes). This article focuses on the type II restriction endonucleases, which represent greater than 90% of the known restriction enzymes and are those most used in recombinant DNA manipulations (see Cloning). For more information on types I, II, and III restriction enzymes, see the article on Restriction-Modification Systems.
The sequences recognized by restriction enzymes are 4-8 base pairs in length and are called restriction sites. The hydrolytic scission occurs 5′ to the phosphate in the phosphodiester bond of each strand, so that 5′ phosphoryl and 3′ hydroxyl termini are produced. The hydrolytic cuts can be made either directly opposite one another or offset from one another along the helix, generating either blunt or cohesive ends, respectively, in the resultant DNA fragments (see Restriction Fragment) (Fig. 1). Within the type II family of enzymes, there are several classes (types IIp, IIw, IIn, and IIt), which are distinguished by the nature of the sequence recognized and by the position of cleavage within or outside of the site (1). Representative examples are given in Table 1.
Figure 1. Types of cleavage by restriction endonucleases and resulting DNA termini: (a) staggered cut resulting in cohesive ends; (b) blunt cut resulting in blunt ends. Arrows indicate position of cleavage, and boldface p indicates phosphodiester bond cleaved.
Table 1. Classes of Type II Restriction Enzymes
a The allowance of one or more nucleotide substitutions at a specific position in the restriction site. b Py is Pyrimidine containing nucleotide residue (C,T), and Pu is purine containing nucleotide residue (A,G).
c Indicates the position of the site of cleavage of the "top" and "bottom" strands 3′ to the recognition site.
d N is G,A,T, or C.
The cuts produced by restriction enzymes yield defined fragments of DNA that can be exploited in several recombinant DNA procedures. Appropriately cleaved fragments of duplex DNA produced from restriction digestion can be ligated (joined) to other fragments using a variety of methods (Table 2) (see also Ligation). For fragments containing compatible cohesive ends (3′ or 5′ overhangs) resulting from digests with the same enzyme, such as EcoRI or Pst I, ligation of both fragments with DNA Ligase regenerate the same restriction site. Fragments produced by different enzymes but having compatible termini can generate novel cleavage specificities on ligation. For example, SalI (G A TCGAC)-digested and AvaI (C A TCGAG)-digested DNA fragments contain 5′ overhangs that are compatible with one another. On ligation of the fragments, both the SalI and AvaI sites are destroyed, and only a TaqI (a TCGA) restriction site remains in the product DNA.
Table 2. Manipulations of DNA Fragment Ends by DNA Ligation
|
DNA fragments |
DNA restriction |
DNA restriction |
Resultant, new |
|
containing: |
fragment 1, with a |
fragment 2, with |
restriction site |
|
a |
upon |
||
|
given Terminus: |
given Terminus: |
ligation of |
|
|
fragments 1 & 2: |
a Italics represent nucleotides added by Klenow fragment of DNA polymerase I.
New cleavage sites can also be generated by ligating blunt-ended termini together (see Blunt-End Ligation). For example, a MboI site (A GATC) can be generated by ligating two blunt-ended fragments: one generated by the blunt-end producer EcoRV (GAT a ATC) and the other by StuI (AGG a CCT). Fragments with incompatible cohesive 5′ ends can be converted to blunt-ended, double-strand molecules with the Klenow fragment of DNA polymerase I. For example, fragments containing the 5′ overhang of an EcoRI site (G a AATTC) can be filled in with Klenow fragment to form blunt-ended molecules with the 5′ sequence AATT and ligated to blunt-ended fragments produced by HaeIII (GG A CC); the product DNA then contains a regenerated EcoRI site (GGAATTC). Fragments containing either 5′ or 3′ overhanging ends can also be made blunt-ended by removing the single-stranded segment with either S1 nuclease or mung bean nuclease and ligated to other blunt-ended termini. Fragments produced by digestion with two different restriction enzymes, for example, with single PstI and BamHI sites, will have different termini (PstI or BamHI) on each end that can be ligated into cut DNA with similar, compatible termini.
Orientation problems are usually found with blunt-end ligations. For example, a gene borne on a blunt-ended DNA fragment ligated into a vector downstream of a promoter has only a 50% chance of being ligated in the proper orientation for expression, because blunt-end fragments can be joined in either orientation. This can be overcome with ligations using cohesive-end or double-digested fragments, which assure correct orientation of the gene on ligation, using restriction enzymes of the type IIp, IIw, IIn, and IIt classes (Table 1) (1).
The type IIs restriction enzymes (see Restriction-Modification Systems) offer unique opportunities for molecular cloning experiments because they cleave outside, but 3′ adjacent to, the sequence recognized, producing long staggered ends (2). One application involves the design of an adapter with a type IIs site within an engineered single-stranded region that anneals to a complementary single-stranded DNA template. This adapter can first be utilized as a primer for DNA polymerase to synthesize a double-stranded template, and this product can subsequently be hydrolyzed with the type IIs restriction enzyme to achieve the desired cut. Another application is for the precise excision of DNA fragments and for gene assembly using a DNA vector containing a unique restriction site immediately between two inward-facing type IIs sequences (2, 3). These applications illustrate why type IIs enzymes have been referred to as "universal" restriction enzymes.
Restriction enzymes can be used in combination with methyltransferases to generate new or rare cleavage specificities (see Methylation, DNA and Methyltransferase). For example, premethylation of (GGATCCCGG) sequences with the methylase M*MspI to form mCCGG inhibits methylation by M* BamHI (GGATmCC), but not cleavage by the restriction enzyme R*BamHI. Therefore, all BamHI sequences not adjacent to methylated MspI sites will be rendered insensitive to BamHI cleavage after methylation by M*BamHI (4). Another application, methylase-limited partial digestion, involves incubation of DNA for limited times with methyltransferases in order to methylate some but not all canonical sequences. Therefore, some of the canonical sequences remain susceptible to cleavage by the partner endonuclease, while others do not. This method is convenient when large restriction fragments are desired (5).
Given the variety of sequence specificities, cleavage and methylation properties of available restriction and methylation enzymes, and the numerous ways in which they can be used in DNA manipulation and analysis, it is clear why the discovery of these enzymes revolutionized molecular cloning. Other applications of these enzymes are discussed in the articles on restriction map and Restriction Fragment.