Most of the proteins made by a cell are retained intracellularly; only a specialized subset is secreted outside the cell. Proteins secreted by cells have several major functions, including signaling to other cells, formation of an insoluble extracellular matrix surrounding the cell, and degradation of extracellular material. Molecular biology has helped explain how newly synthesized proteins are selected for secretion and transportation across the hydrophobic barrier of the plasma membrane (see Protein biosynthesis).
The mechanism for transporting newly synthesized proteins is highly conserved from bacteria to mammals. A key difference, however, is that bacteria translocate the proteins directly across the plasma membrane to the outside world, whereas eukaryotic cells translocate them into a specialized intracellular organelle, the endoplasmic reticulum. Newly synthesized proteins are conveyed from the endoplasmic reticulum to the cell surface via a series of carrier vesicles. It is therefore useful to consider prokaryotic and eukaryotic protein secretion separately.
1. Protein Export from Bacteria
Bacteria are extremely simple from a cell biological point of view. Gram-positive bacteria have only a single membrane, the plasma membrane, separating their cytoplasm from the outside world. Gram-negative bacteria have a second outer membrane, separated from the inner membrane by periplasmic space. In this case, the newly synthesized protein that is to be exported must cross the outer membrane in addition to the inner plasma membrane.
The basic elements of translocation across the membrane appear to be universal (1). A small pore or hole through which unfolded proteins can pass traverses the membrane. Energy is used to push unfolded protein through the hole. Molecular chaperone proteins exist on the cytoplasmic side of the pore to maintain a protein in an unfolded state, which allows it to be threaded through the pore. The chaperones distinguish proteins that are to be exported from cytoplasmic proteins because the exported proteins have signal sequences, regions of contiguous amino acids that are usually at the amino terminus of the protein. Once the protein has crossed the membrane, additional chaperones can facilitate correct protein folding.
Much of the research on protein export by bacteria has utilized Escherichia coli, a gram-negative bacterium. In E. coli there are at least two different ATP-driven transport systems, one that uses the sec A system (see Sec Mutants/Proteins) to pump proteins into the periplasmic space (2), and a second that uses a member of the ABC (ATP-binding cassette) family of proteins to export proteins across both E. coli membranes simultaneously (3).
1.1. The secA System of Protein Transport
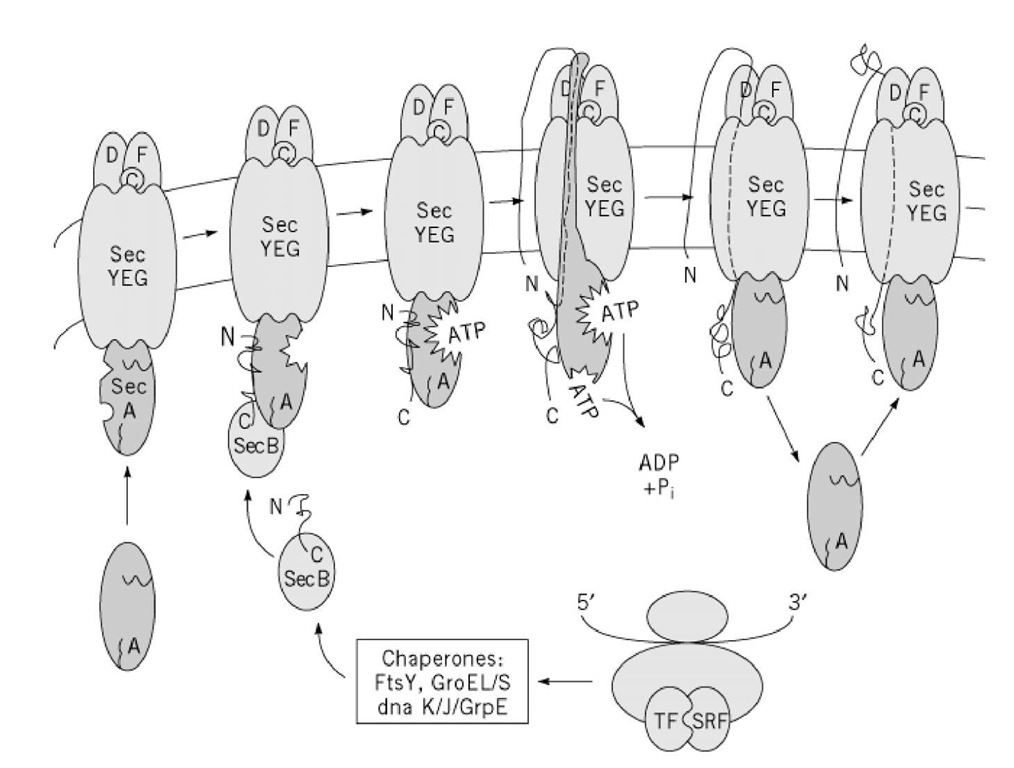
When E. coli makes a protein that is destined for export, it is made as a preprotein containing a signal sequence (4). In most proteins, these signal sequences are a stretch of contiguous amino acids at the amino terminus of a protein, with a positively charged amino terminus, at least six sequential hydrophobic amino acids, and a cleavage site for the signal peptidase that removes the signal after translocation. The presence of the signal sequence in a newly synthesized protein causes a protein chaperone to bind to it, retarding its folding. There appear to be at least two major chaperone systems in E. coli. One involves the tetrameric cytosolic protein secB, which binds to newly synthesized proteins that contain a signal sequence. It binds throughout the length of the newly synthesized polypeptide chain, keeping it from adopting its native conformation (2). A different class of chaperones binds to the nascent chain as the secreted protein is being synthesized on the ribosome. An example of such a chaperone is a ribonucleoprotein, the signal recognition protein (SRP). In E. coli SRP consists of a 4.5 S RNA molecule and a single protein, Ffh, which is a GTPase; that is, it hydrolyzes guanosine triphosphate (5).
The proteins destined for export, complexed with chaperones that keep them from folding, interact with receptors on the bacterial plasma membrane to initiate translocation across the membrane. The secB receptor is secA, and the SRP receptor is another GTPase, FTsY (5). The signal sequence has therefore two functions: (1) attracting a chaperone and (2) helping to target the complex to plasma membrane receptors.
The actual translocation event is through a transmembrane pore. In E. coli the pore is made up of the sec YEG complex, consisting of three major membrane proteins (secY, secE, and secG) with several accessory proteins. The sec YEG complex forms the pore through which unfolded proteins pass on their way out of the cell. Translocation involves pushing the unfolded peptide through the pore by a mechanism that resembles a sewing machine needle or a piston (6). A peripheral membrane protein (secA) takes a 25-residue loop, formed by the amino-terminal signal peptide and the contiguous amino acids, and pushes the loop through the membrane. The amino-terminal amino acids remain cytoplasmic. To do this, the secA protein goes through a remarkable, ATP-driven, conformational change, which causes an arm of the secA protein to be inserted transiently into and partially across the membrane. After the pushing step, ATP is hydrolyzed and secA is returned to a cytoplasmic location, ready for a second translocation pumping cycle. The signal peptide is cleaved off by a "signal peptidase" located in the periplasmic space. Further translocation in 25-residue steps can be driven by additional cycles of protein activity. After the initial step, however, further translocation is facilitated by a transmembrane proton motive force (Fig. 1).
Figure 1. The SecA system of protein transport. Chaperones bind to a nascent protein in order to keep it an unfolded stal it to receptor SecA at the membrane. SecA translocates the protein across the membrane through the protein pore (SecYl conformational changes.
Bacteria may push protein across the translocation channel, rather than pull it, because the source of energy, ATP, is exclusively cytoplasmic. Once in the periplasmic space, secreted proteins form disulfide bonds and fold into proteinase-resistant conformations. Such folding is necessary before the proteins are transported through holes, poorly described, in the outer membrane.
1.2. ABC Protein-Mediated Export
Some proteins destined for export are recognized in the cytoplasm of E. coli by an entirely different class of signal sequences. In these cases, the sequence is at the C-terminus and is not cleaved off by a signal peptidase after transport. Proteins with this type of signal sequence are closely related to each other and include such proteins as toxins, proteinases, and lipases. The signal sequence is recognized by a different class of plasma membrane protein, ATP-driven protein translocators of the ATP-binding cassette (ABC) family (3, 7). ABC proteins have two cytoplasmic ATP-binding domains and two hydrophobic domains, with six transmembrane sequences. They can be either a single polypeptide or be made up of several polypeptides. A complex of a bacterial ABC export with two accessory proteins allows it to export a cytoplasmic protein across both the inner and outer bacterial membrane at the same time. Assembly of the transporting complex is triggered by substrate binding (8).
2. Protein Export from Eukaryotes
Unlike bacteria, eukaryotic cells are packed with intracellular membranes. A considerable fraction of those membranes is involved in protein export. Thus protein export in eukaryotes involves translocation across membranes but also transport of the newly synthesized protein through intracellular compartments (9). Protein export in eukaryotes is thus a much more complex process than in bacteria. It becomes necessary to know the intracellular pathway taken by a newly synthesized protein before it reaches the eukaryotic cell surface (see also Protein targeting).
2.1. Intracellular Trafficking of Exported Protein
For the most part, eukaryotes do not secrete proteins directly across the plasma membrane. Instead, the newly synthesized proteins are translocated from the cytoplasm into an intracellular organelle, the endoplasmic reticulum (ER). The ER is a series of membranous cisternae and tubules that spread throughout the cytoplasm and is continuous with nuclear membrane. As we shall see, the mechanisms for translocating proteins across ER membranes and bacterial membranes both involve a transmembrane pore. Why have an ER? A common speculation on the evolutionary advantage of the ER is that it provides a controlled milieu in which exported proteins can fold and oligomerize, without being exposed to the rigors of the extracellular world. In this regard, the lumen of the ER resembles the periplasmic space in gram-negative bacteria.
A protein, correctly folded and oligomerized in the ER, must be transported to the outside of the cell. The initial step in transport from the ER is the formation of small membrane vesicles, usually called carrier vesicles, that contain the exported proteins as cargo (10, 11). Formation of carrier vesicles occurs at specialized ER regions called transitional zones, and it requires a cytoplasmic coat, to impart curvature to the vesicle membrane and to pinch it off from the donor ER (12).
After a carrier vesicle is formed, it must recognize and fuse with its target. Recognition and fusion (see Exocytosis) involve proteins on the vesicle (v-SNARE) and on the target membrane (t-SNARE) (13). When a carrier vesicle leaves the ER, it does not go directly to the plasma membrane but instead fuses with an organelle, the Golgi complex, which is a mandatory way station on the secretory pathway of eukaryotes. The Golgi complex is a stack of membranous cisternae, similar morphologically to a stack of pancakes. The carrier vesicles enter the cis end of a Golgi stack and exit from the trans side. A protein to be exported goes through a series of glycosylation steps in which six- or nine-carbon sugars are added to or removed from oligosaccharide chains attached to serine, threonine (O-glycosylation), or asparagine residues (N-glycosylation). The added sugars can often protect the exported protein from rapid proteolytic degradation after it is secreted into the extracellular world. A unique class of oligosaccharides is added to newly synthesized lysosomal enzymes that allows them to be diverted out of the secretory pathway to primary lysosomes.
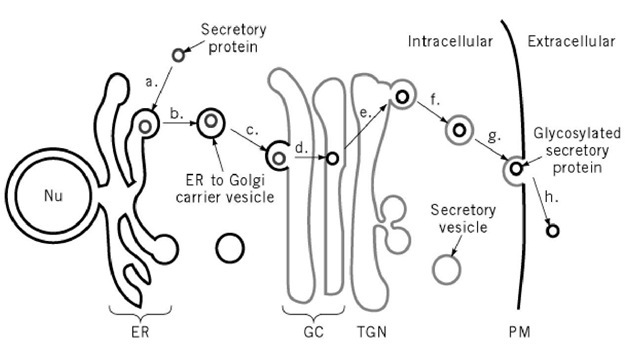
A second budding event takes place from the trans region of the Golgi complex. A second coating mechanism causes the formation of a secretory vesicle containing glycosylated proteins for export. The post-Golgi secretory vesicles move to the plasma membrane, either by diffusion or by transport along microtubules. When the secretory vesicles reach the plasma membrane, their membranes and the plasma membrane fuse to form one continuous bilayer, a process known as exocytosis. The fusion step results in the release of the exported protein into the extracellular medium (Fig. 2).
Figure 2. The eukaryotic secretory pathway. Secretory proteins enter the membrane trafficking system by (a) translocation from the cytoplasm into the endoplasmic reticulum (ER) (b) In the ER, proteins are packaged into ER to Golgi complex (GC) carrier vesicles that (c) deliver proteins to the GC. As proteins progress through the GC they become (d) glycosylated. Once the (e) reach the /rans-Golgi network (TGN), they (f) are sorted into secretory vesicles in which proteins are (g) transported to the plasma membrane (PM). At the cell surface secretory proteins are (h) released into the extracellular environment.
This pathway of protein export or secretion is the one followed in most organisms, including yeast, protozoa, and mammalian cells such as muscle cells and fibroblasts. It is commonly referred to as the constitutive pathway, to distinguish it from a specialized pathway found in specialized cells, such as endocrine and exocrine cells, and in cells of the hematopoietic system, such as neutrophils or cytotoxic T lymphocytes . In such cells, newly synthesized proteins are diverted out of the normal biosynthetic pathway to be stored in secretory granules (14). Fusion of the secretory granule with the plasma membrane is usually triggered by an external stimulus. Because export of this class of secreted proteins is controlled by a stimulus, the triggered release of protein from a storage pathway has been called regulated release.
The protein export pathway of eukaryotes is highly conserved. Protozoa, yeast, and mammalian cells use essentially the same mechanisms to translocate proteins into an ER, to glycosylate and sort them in the Golgi, and to export them across the plasma membrane by exocytosis.
2.2. Translocation Across the ER Membrane
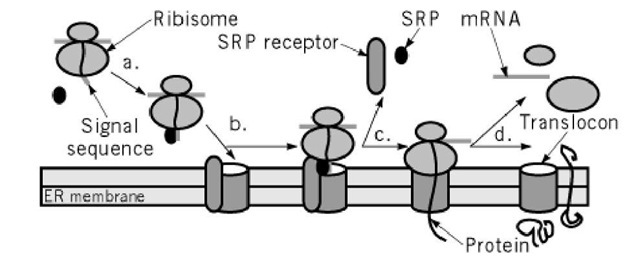
Translocation of newly synthesized proteins across the ER membrane shows many similarities to translocation across the plasma membrane protein of bacteria (1, 15, 16). Proteins are prevented from folding in the cytoplasm. They are fed across the plasma membrane through a translocon, a proteinaceous pore, which has three subunits very similar to the bacterial proteins made by the secY, E, and G genes. By electron microscopy, these pores are rings about 8 to 10 nm in diameter, with a central pore of 2 nm, sufficient to allow the passage of an extended, hydrated peptide of 1.1 nm in diameter. These pores can now be recognized (17). In yeast, proteins traverse pores in the ER by two different types of translocation mechanisms. One is an ATP-driven process that translocates proteins whose synthesis is complete. The other couples translation to the translocation process. In this transport mode, the ribosome is attached to the proteinaceous transport pore, the translocon, and feeds the nascent train through the pore as it is being synthesized. Mammalian cells only have the co-translation made of translocation. When translocation is co-translational, the nascent chain is recognized in the cytoplasm by a signal recognition particle, which stops further protein synthesis until the complex of ribosome, nascent chain, and signal recognition particle reaches the endoplasmic reticulum (Fig. 3).
Figure 3. Translocation across the ER membrane. (a) Signal recognition particle (SRP) binds to a signal sequence on a nascent protein and halts the protein’s synthesis by the ribosome. (b) SRP brings the protein and ribosome to the translocon on the ER membrane by binding to its receptor. (c) Protein synthesis restarts when the ribosome is correctly positioned and the protein has been threaded into the translocon. (d) Proteins can either be translocated completely into the lumen of the ER to become secretory proteins or translocated partially through the membrane to become integral membrane proteins.
As the nascent chain enters the cytoplasm, it is glycosylated on suitable asparagine residues and forms disulfide bonds. Folding is completed by association with accessory proteins such as BiP, calnexin, peptidyl prolyl cis/trans isomerases, and protein disulfide isomerase (18). Proteins also oligomerize in the lumen of the ER. For example, protocollagens trimerize with an extended coiled-coil configuration, accompanied by cross linking between hydroxyproline and hydroxylysine residues (see Hydroxylation (Lysine, Proline)). If cross linking of collagen protofibrils is inhibited, the improperly folded collagen cannot leave the ER and is quickly degraded. A similar observation is made with the secreted protein al-antitrypsin. In the disease, a-antitrypsin deficiency, a mutation in the protein can prevent its correct folding. These two examples demonstrate that there is a quality-control system that surveys the proteins leaving the ER and prevents the export of improperly folded proteins. Recent data suggest that improperly folded ER proteins are exported from the ER back to the cytoplasm, where they are degraded by proteasomes (19).
2.3. Traffic of Secretory Proteins from the ER
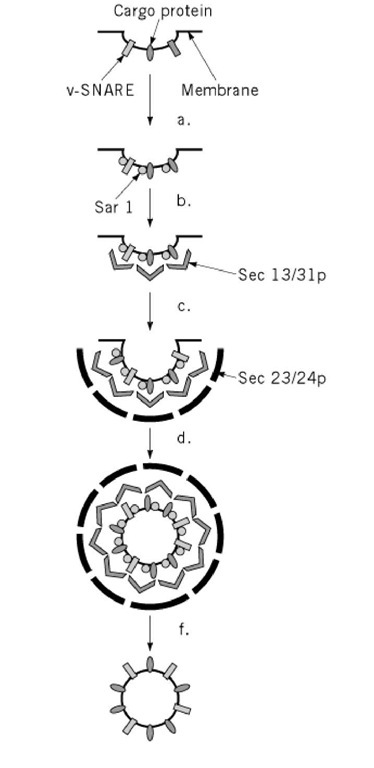
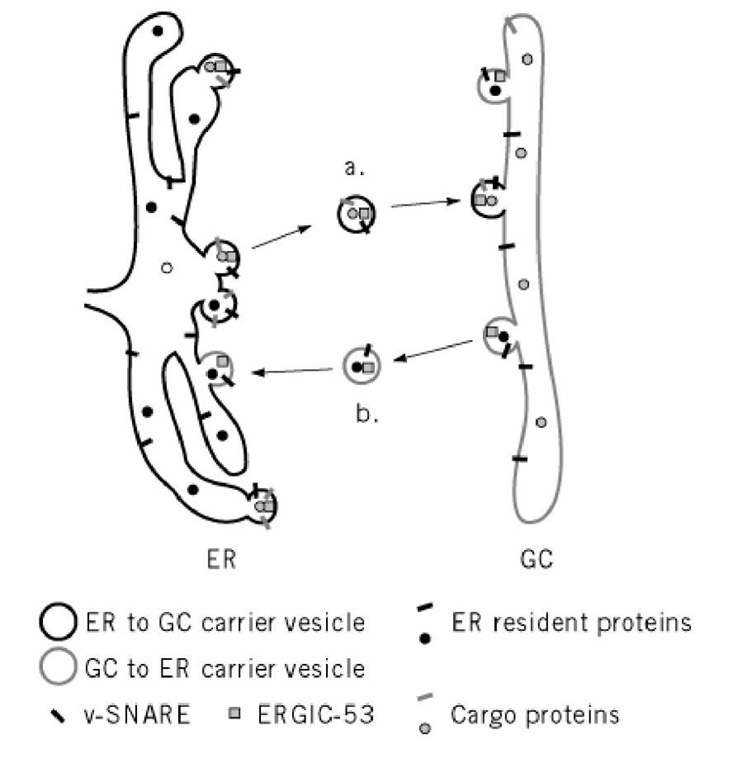
To take correctly folded and oligomerized proteins from the ER, a vesicle forms in the transitional elements and includes proteins to be exported, but excludes resident proteins of the ER lumen, such as BiP (20). The coat that causes the vesicle to form is now known as COPII. Yeast COPII contains four subunits, sec31p, sec13p, sec23p, and sec24p (see sec mutants). Assembly of a COPII coat requires a small GTPase, Sar1, and a guanine nucleotide exchange factor, Sec12p, in the ER membrane (12) (Fig. 4). The coated vesicle leaving the Golgi carries with it a complement of v-SNARE molecules (see Exocytosis) to allow it to fuse with the cis-Golgi network. In yeast, these are Sec22p, Bos1p, and Bet1p. Resident proteins such as BiP may be excluded from the lumen of the coated vesicle because they are oligomerized into complexes that are too big to enter the small vesicle. To some extent, exported proteins are those that lack a retention signal and so are not retained in the ER. Export of secreted proteins would then be by default, because they lack information to go anywhere else. There is evidence, however, that positive sorting occurs (21) (Fig. 5). In yeast, the secreted protein invertase is recognized by a membrane-bound ER protein (Emp24p) that is required for its transport to the Golgi (22). Furthermore, cargo proteins are concentrated as they leave the ER (23, 24). Since most soluble resident proteins in the ER lumen are not glycosylated, an attractive hypothesis is that exported proteins are recognized by a lectin, which concentrates them in budding vesicles. A protein, ERGIC-53, recycles between the ER and the Golgi and is a lectin with the capacity to bind the mannose residues found on newly synthesized secretory proteins (25). Proteins such as ERGIC-53 might bind secreted proteins and actively carry them to the Golgi complex, in the same way that the mannose phosphate receptor carries newly formed lysosomal enzymes to the prelysosomal compartment.
Figure 4. Formation of ER to GC carrier vesicles. ER to GC carrier vesicles are formed with the COPII coat (Sar1, Sec 13/31p, and Sec 23/24p). Components are thought to bind sequentially with (a) the GTPase Sar1 (in its GTP state), binding to v-SNARE and cargo molecules. (b) Sec 13/31p binds to Sar1 and then (c) Sec 23/24p binds to the whole complex. By unknown mechanisms the COPII coat (d) causes a vesicle to bud off from the ER membrane, and (e) uncoating leads to the (f) mature carrier vesicle, which can then go on to fuse with the Golgi complex. Formation of other types of vesicles is likely to involve similar steps using different coating proteins.
Figure 5. Sorting of proteins between the ER and GC. (a) Cargo proteins destined for secretion and v-SNAREs needed for carrier vesicle fusion to the GC are sorted away from the ER resident proteins into COPII coated vesicles. After delivery to the GC, the cargo proteins continue through the secretory pathway. (b) However, v-SNARES needed for the ER to GC trafficking step and ER resident proteins that have been mistakenly transported to the GC can be retrieved from the secretory pathway through retrograde transport back to the ER by COPI-coated carrier vesicle. ERGIC-53, a likely carrier protein, cycles between the ER and GC and may assist in sorting the secretory proteins from proteins that are retained in the ER.
Proteins such as ERGIC-53 and the v-SNARE proteins can recycle back to the ER from the Golgi. The vesicle for Golgi to ER flow is made up of coatomers, referred to as COPI coatomers. Coatomers consist of seven subunits. The small GTPase active during coatomer coat formation is an ADP ribosylation factor (ARF1).
Fusion of the ER-derived carrier vesicle with the Golgi membranes requires the v-SNAREs Sec22p, Bos1p, and Bet1p in the vesicle membrane, and the cognate cis-Golgi t-SNARE, the sed5 protein. In addition, a second class of small GTPases are required that are members of the rab family (26). The rab GTPases are ras-like proteins that are specific to a certain organelle or a certain trafficking step. In yeast, mutations in the rab protein ypt1 inhibit fusion with the Golgi and cause the accumulation of carrier vesicles. The ypt1 protein appears to play a role in v-SNARE/t-SNARE interactions (see Exocytosis) involved in cis-Golgi targeting. Two of the v-SNAREs in yeast, Bos1p and Sec22p, normally form a complex. The complex does not form if ypt1 is absent, nor is a v-SNARE/t-SNARE complex formed, suggesting that ypt1 plays a role in activating the v-SNAREs, allowing a v-SNARE/t-SNARE complex to form. It is also possible, however, that ypt1 is also activating the t-SNARE complex on the target membrane, by removing its chaperone, Sly1p. This chaperone is a member of the Sec 1p family of syntaxin-family Chaperones (see Exocytosis).
2.4. Modification of Secreted Proteins in the Golgi
As secreted proteins pass through the Golgi complex, they can be modified by glycosylation, sulfation, or proteolysis. Three major types of glycosylation occur, N-linked, O-linked, and glycosaminoglycan addition. In mammalian cells, the mannose-rich, asparagine-linked oligosaccharides that had been added in the ER are first trimmed in the Golgi complex and then rebuilt. Mannoses are first removed and then replaced by other sugars. On a stump of the original mannose tree, a multichain oligosaccharide is generated by the orderly addition of the sugars N-acetylglucosamine, galactose, and sialic acid (N-acetylneuraminic acid). The previous form of glycosylation is referred to as N-glycosylation, since the sugars are added to an asparagine residue. If a sugar chain is added to a serine or threonine residue, it is referred to as O-glycosylation. The first sugar added is usually N-acetylgalactosamine. The oligosaccharide chains are branched but are usually shorter and more variable than N-linked oligosaccharides.
The third type of glycosylation results in the formation of proteoglycans. Proteoglycans are formed by the addition of xylose to serine residues as the protein moves from the ER to the Golgi (27), followed by the addition of a long, highly charged, unbranched oligosaccharide called a glycosaminoglycan. Glycosaminoglycans are polymers of a disaccharide unit, one of which is usually uronic acid. The other member of the oligosaccharide pair varies, depending on the type of glycosaminoglycan. Part of the strong negative charge on the proteoglycans is due to the addition of sulfate groups to the glycosaminoglycans. Sulfate can also be added directly to tyrosine residues on secreted proteins. Sulfation of proteins only takes place in the late Golgi. Radioactive sulfate is thus commonly used to label exported proteins selectively and to identify a set of proteins at a late Golgi step of transport. Glycosaminoglycan chain synthesis can be initiated in the late Golgi by growing cells in the presence of a membrane-permeable derivation of xylose (28). This technique has also been used to identify membrane traffic from the late Golgi to the surface of the cell.
Several proteins, membrane associated and secreted, are subject to proteolytic processing as they pass through the Golgi complex. The proteolysis is highly specific, occurring usually to the carboxyl side of a four-residue consensus sequence: Arg-X-(Lys/Arg)-Arg (29). The Golgi enzyme that carries out this proteolysis is a membrane-attached form of the subtilisin family, called furin. Secreted proteins cleaved by furin include the hepatocyte and nerve growth factors and pro-albumin.
Exported proteins can take one of two pathways to leave the Golgi complex, the constitutive and the regulated [Fig. 4 in (14)]. In the regulated pathway, newly synthesized proteins are stored in specialized secretory granules until cells receive a signal to secrete. Storage does not occur in the constitutive pathway; nascent proteins go directly from the Golgi to the cell surface. Little is known about sorting into constitutive carrier vesicles, the coat required to generate a carrier vesicle, or the v-SNAREs and t-SNAREs required for fusion with the cell membrane. Much more is known about the regulated secretory pathway, which packages newly synthesized proteins into secretory granules (qv). Similarly, study of the regulated secretory pathways has revealed a great deal about the final step, fusion with the plasma membrane, exocytosis.
2.5. Nonclassical Protein Secretion
The secA-mediated pathway of bacterial protein export uses a translocon or transmembrane pore that is closely similar to the ER translocon in eukaryotes. There is also a eukaryote equivalent of the ABC transporter system in bacteria. The yeast protein, Ste6p, is an ABC protein that exports a-factor, a small yeast mating-type peptide, from the cells. There is no evidence, however, that eukaryotic ABC proteins can translocate full-length proteins across the plasma membrane.
Yeast can export proteins without using the classical, ER to Golgi pathway. Mutations in the classical pathway that block invertase secretion have no effect on this pathway, called the nonclassical pathway. The gene for a transmembrane protein, nce2, that is important for nonclassical secretion has been cloned and sequenced, but its role in protein export remains unclear (30).
Mammalian cells export proteins, such as interleukin-1b, basic fibroblast growth factor, and thioredoxin, that do not have a classical signal sequence (31). In addition, drugs such as monensin and brefeldin A that normally block classical secretion are without effect on these proteins. Their secretion has been linked to the heat-shock response. Little is currently known about the molecular mechanisms that are involved.