The parasexual cycle allows new allele combinations to be formed independently of whether the organism can undergo a sexual cycle and meiosis. The parasexual cycle involves the fusion of haploid nuclei, mitotic recombination, and random chromosome losses. Parasexuality was discovered by Giorgio Pontecorvo (1) and has been demonstrated with several fungi and, in a modified form, with cultured cells of multicellular animals.
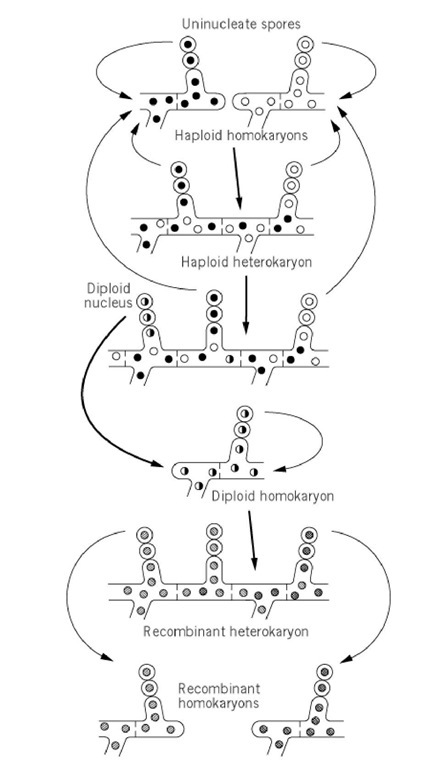
The parasexual cycle is sketched in Fig. 1, taking a filamentous Ascomycete as a model. Fusion of haploid cells to form a heterokaryon occurs spontaneously in many fungi in a process called hyphal anastomosis. In many fungi this process occurs with certain pairs of strains said to be vegetatively compatible and fails with others. Compatibility requires the strains to carry certain combinations of alleles at certain genes. Heterokaryons can be prepared in the laboratory by fusion of protoplasts and other techniques in fungi without anastomosis and in many other organisms.
Figure 1. Parasexual cycle in filamentous Ascomycetes . Nuclei are followed from the haploid parents above to the haploid segregants below, through cell fusion, nuclear fusion, and haploidization. Recombinant nuclei are indicated by different shading patterns.
Two haploid nuclei in a heterokaryon may fuse to form a diploid nucleus. This process occurs seldom or not at all in vegetative heterokaryons, although it may be very quick after the fusion of gametes in the sexual cycle. Diploid cells are usually very hard to distinguish from heterokaryotic cells carrying the same genetic markers, but they may be very easily distinguished from haploid cells. In fungi in which all spores are uninucleate, colonies grown from diploid and haploid spores are distinguished easily with the help of appropriate genetic markers.
The diploid nuclei of many fungi are unstable because the loss of one chromosome of each pair is not deleterious. A slow process of haploidization takes place during mitotic growth as random chromosome losses convert the diploid into haploids through intermediates called aneuploids. The resulting haploid nuclei contain random combinations of the chromosomes of the two original haploid nuclei. The frequency of chromosome loss is increased by treatments with benomyl and other agents.
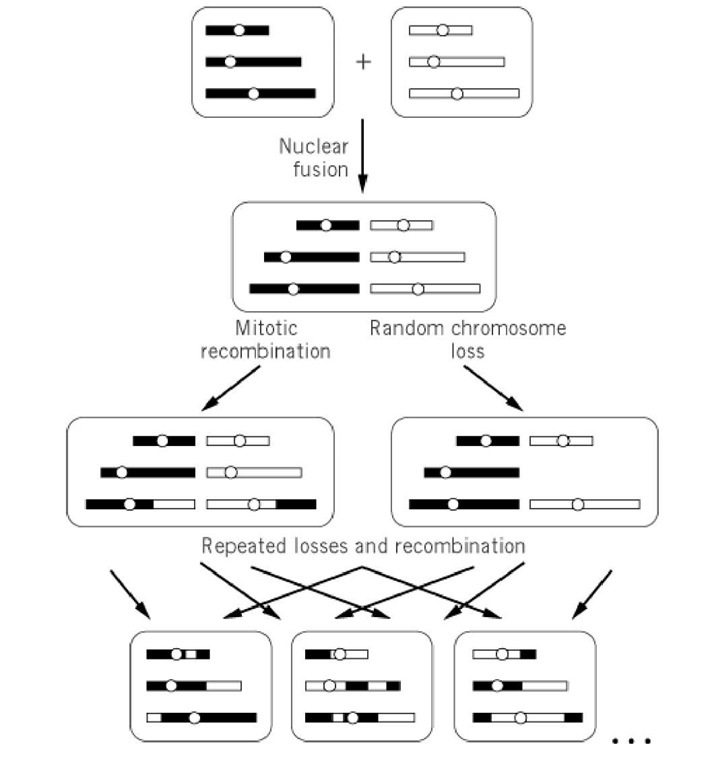
In addition, mitotic recombination occurs sporadically between the duplicated chromosomes of the diploid and the aneuploids and produces new combinations of their genetic markers (see Mitotic Recombination). The chromosomes in the final haploid nuclei are composed of random combinations of chromosome segments from the two original haploid nuclei (Fig. 2).
Figure 2. Chromosome events during the parasexual cycle. Three chromosomes are followed from the haploid parents above to the haploid segregants below, through diploidization, mitotic recombination, and chromosome loss.
As a result of the parasexual cycle, genes in different chromosomes exhibit recombination frequencies of 50 % while those that are close to each other in the same chromosome appear more or less tightly linked.
Fusion of cultured cells from different mammals gives rise first to heterokaryons and then to anfiploid nuclei, which contain all the chromosomes of the original cells. Chromosome losses reduce the number of chromosomes in these anfiploids, but not necessarily at random: chromosomes of Homo sapiens are lost more frequently than those of Mus musculus from their anfiploids. The correlation between the presence or absence of certain genetic markers and certain chromosomes in the segregants is a powerful technique for the quick assignment of genes to chromosomes (see Cell fusion, Hybridomas).