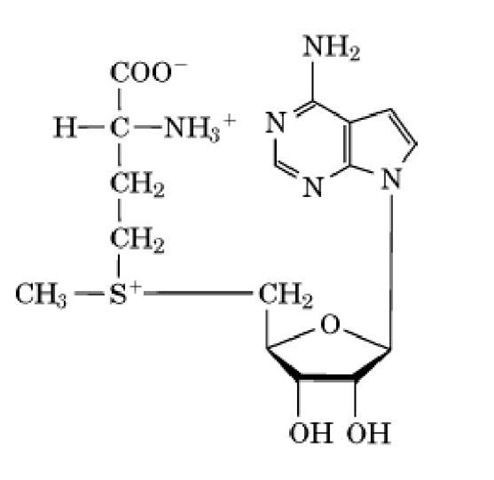
Methyltransferases use -adenosylmethionine (AdoMet) as a methyl donor to catalyze methylation of functional groups on amino acids, usually on their side chains (1). Many different amino acids in proteins are methylated by a host of specific enzymes (see Methylation, Protein). Several instances of these are described in this article. The methyl group donated from AdoMet is on a sulfonium ion (trivalent sulfur atom having a "formal" positive charge) (Fig. 1) and is readily and irreversibly transferred to other atoms (1). AdoMet is an "expensive" molecule because it is synthesized from methionine and ATP with production of inorganic phosphate, P^ and pyrophosphate, PP^; the latter is rapidly cleaved to 2 Pj by pyrophosphatases. The net cost is three "high-energy" bonds (see Adenylate Charge), which signifies that maintaining AdoMet levels is a high priority for cells generally, apparently to methylate proteins and DNA (1).
Figure 1. Structure of S-adenosylmethionine.
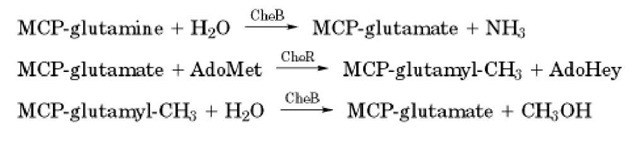
Perhaps the most well characterized methyltransferases are the CheR methyltransferases in bacteria involved in Chemotaxis. These enzymes catalyze the methylesterification of a glutamate (E) residue within the protein consensus sequence (A/S)XXEEX(A/T/S)A(A/T/S) (2). These residues are located on two a-helical coiled coils within the cytoplasmic region of the receptors, known as methyl-accepting chemotaxis proteins (2). These methylated residues are located in different places in the receptors of Bacillus subtilis and Escherichia coli (3). In E. coli the effect of methylation is to increase kinase activity. In B. subtilis, the position of the site of methylation governs whether CheA kinase activity is increased or decreased. Interestingly, in some instances the glutamate residue is produced from a genetically encoded glutamine (see Genetic Code) by the CheB methylesterase acting as a deamidase. This enzyme also hydrolyzes the methyl esters produced by the action of CheR (2) (Fig. 2).
Figure 2. Reactions catalyzed by CheR methyltransferase and CheB methylesterase.
In contrast with CheR, the CheB methylesterase of both E. coli and B. subtilis is activated by phosphoryl transfer from phosphorylated CheA and, hence, becomes stimulated when CheA is activated (2, 4). After autophosphorylation, the CheA kinase leads to increases in the concentrations of the phosphorylated forms of CheY and CheB. The former enhances tumbling in E. coli and smooth swimming in B. subtilis, whereas the latter causes demethylation of the receptors to reduce the activity of the CheA kinase (4). In other words, upon activation, CheA simultaneously causes an excitatory event (due to phosphorylated CheY) and sets in motion the adaptation process (due to the action of phosphorylated CheB). Moreover, in B. subtilis, both CheR and CheB are much more active immediately after the addition or removal of attractant rather than later, probably due to slow conformational changes in the receptors exposing the sites of methylation.
Other methyltransferases catalyzing methylesterifications have been identified and characterized. These include the enzyme that catalyzes the metabolically labile methylation of the carboxyl-terminal leucine residue of protein phosphatase 2A, an enzyme important in regulating cell metabolism (7). The methylation and demethylation represent a novel device for regulating the phosphatase itself. The membrane-bound methyltransferase that catalyzes methylation of carboxylterminal isoprenylcysteine residues, which in yeast is the product of the STE14 gene, has also been characterized, and strains lacking it have been studied (see Prenylation). Among other consequences, the a-mating type factor activity is much reduced (7).
In bacteria, the methyltransferase that catalyzes methylation of isoaspartate residues, an isomer of aspartate that occurs when proteins age, has been identified and the gene disrupted. This enzyme helps with reisomerizing isoaspartate back to aspartate, restoring the original protein. Bacteria mutant in this gene are sensitive to heat shock and survive poorly in a stationary phase, results that imply the importance of this type of protein repair (7).
Arginine methylation in eukaryotes is performed by a specific subfamily of methyltransferases. This family of methyltransferases contains AdoMet binding motif similar to small molecule and nucleic acid methyltransferases (8). Many of the methyltransferases also include a C-terminal domain involved in arginine substrate recognition. The typical recognition motif for the methyltransferase is RGG, RXR, and RG, which can be methylated in three ways: N -monomethylarginine (MMA), N N (asymmetric) dimethylarginine (aDMA), and N N (symetric) dimethylarginine (sDMA).
Methylation of these conserved sites is also selective. RGG recognition sites have only been isolated as MMA and aDMA and no sDMA, while RXR and RG motifs have been isolated as aDMA- methylated but not MMA or sDMA. SmD1 and SmD3, spliceosomal small nuclear RNA binding proteins, have been isolated with sDMA methylation on the site GRG leading to the possibility that more recognition motifs exist and provide specificity as well as control of extent of methylation by the arginine methyltransferase.
Histone methylation at specific N-terminal lysine residues is believed to be associated with the epigenic inheritance of heterochromatin (transcritionally silent DNA), and transcriptional regulation (9). This modification is catalyzed by histone methyltransferases (HMT) on K4, K9, K27, and K36 of histone H3 and K20 of histone H4 in many organisms, although the pattern may be species-specific (9). Like all methyltransferases, HMT contains an AdoMet-binding motif, and, as arginine’s guanidino group is multiply methylated, a single lysine can be mono, di, and tri-substituted. Little information is available concerning HMT regulation, although, in developing rat neuron HMT, activity is enhanced, and adult rat neuronal HMT activity is reduced (9).