From an incubation of waste water samples with several types of Escherichia coli bacteria, Hofschneider (1) discovered in 1963 some new types of bacteriophages, among which was one designated M(unich)13. Since then, a large number of bacteriophages that constitute the family of filamentous phages (Inoviridae) have been isolated from different gram-negative bacteria. Filamentous phages belong to the smallest bacteriophages known and differ from most other bacterial viruses in that they are being reproduced continuously upon infection without lysis of their host cells; infected cells continue to grow and divide, although with a reduced rate.
Filamentous phage efficiently infect only host cells that express (sex) pili, which serve as receptor sites. Based on their pilus specificity, different classes of evolutionary closely related filamentous phages can be distinguished. F-specific filamentous phages (the Ff group) only infect male E. coli cells carrying sex pili, encoded by an F factor (ie, a fertility or sex episome). Among these Ff bacteriophages, the almost identical strains M13, fd, and f1 are the most extensively studied and, together with their E. coli host strains, the best characterized from biochemical, genetical, molecular biological, and biophysical points of view.
Because of its relative simplicity and the ease of being genetically manipulated, M13 bacteriophage has a very important role in investigating biological processes that extends far beyond its own life cycle. Apart from the still fruitful investigation of the phage structure and the studies of the processes involved in infection, replication, and assembly of the phage machinery itself, the practical use of M13 phage-derived plasmids as tools in biotechnology is especially enormous (2). The advantages of the M13 phage-derived plasmids as cloning vectors (eg, M13mp18), with their ability to generate large quantities of single-stranded DNA molecules carrying foreign DNA, should be emphasized: Single-stranded DNAs are the templates of choice for DNA sequencing, labeling, or site-directed mutagenesis (3). Furthermore, the phage plays an important role in displaying foreign peptides on the phage surface, when foreign DNA is fused in-frame to a gene coding for one of the coat proteins (see Phage Display Libraries). This technique enables the construction of extensive libraries that can be easily screened to select peptides with specific affinities or activities (4).
1. M13 Phage Particle Architecture
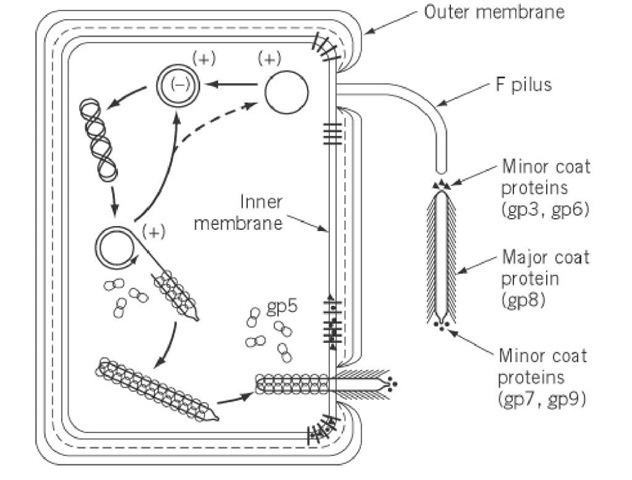
As a member of the filamentous phage family, the M13 phage nucleoprotein particle is long (900 nm) and thin (6.5 nm). The filament contains the viral genome: a twisted closed circular single-stranded DNA molecule of 6407 nucleotides. The DNA genome is protected by a long cylindrical protein coat, consisting predominantly of about 2700 copies of the major coat protein (gp8) and capped at the ends with about five copies each of four different minor coat proteins (5).
The strong native architecture of the phage coat seems to be maintained primarily by hydrophobic interactions between the individual coat proteins. The major coat proteins form a tube around the viral DNA, in an overlapping helical array, with a flexible amphipathic N-terminus located at the outside of the coat and the basic C-terminus interacting with the DNA at the inside of the coat. The hydrophobic domain of the major coat protein is located in the central section of the protein sequence, and it interlocks the coat protein with its neighboring coat proteins (6). As a result, M13 phage is very stable and resistant to most proteinases, salts, extreme pH values, heating, (phospho) lipids, and detergents (but not the strong anionic detergent SDS). The phage particle is sensitive to mechanical stress (ultrasonication) and to chloroform, which in turn makes it susceptible to detergents (7, 8).
The packing of the DNA in the protein tube is determined by an electrostatic interaction between phosphate groups of the DNA and lysine residues of the major coat protein. Elongating the viral genome by adding foreign DNA to the intergenic region ( (3), or reducing the number of positively charged lysine residues of the DNA-binding part of the major coat protein (9), simply results in elongated phage filaments.
The end of the phage that emerges first from the host cell contains a packaging signal as an imperfect double-stranded DNA hairpin loop of the intergenic region. This end of the phage is capped by the minor coat proteins gp7 and gp9 (five copies of each). Both gp7 and gp9 are required for proper initiation of the phage assembly process. At the other end of the phage, five copies each of the other minor coat proteins gp3 and gp6 are located. These proteins are required for properly terminating the assembly process and contribute to phage particle stability (10). The minor coat protein gp3 also contains functional domains enabling host-specific adsorption and subsequent penetration of the viral genome during a next round of infection.
2. Genetic Organization
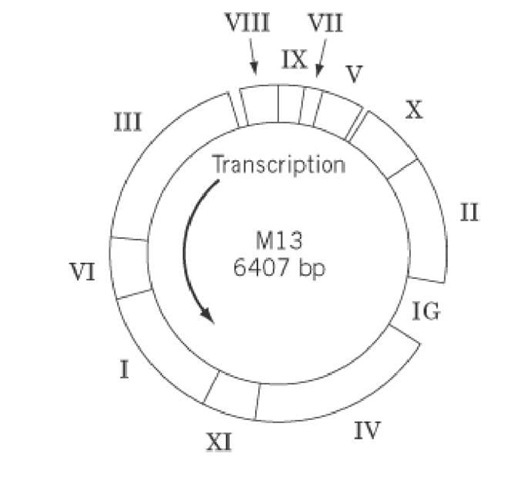
The complete DNA sequence of M13 and related phages is known (11, 12). The genome comprises nine open reading frames, coding for 11 identified proteins (Fig. 1), and two intergenic regions. It should be noted that genes X and XI are contained completely within genes II and I, respectively. The phage-encoded proteins are organized into three functional clusters: gp2, gp10, and gp5 are involved in DNA replication; the morphogenetic gp1, gp4, and gp11 are required for phage assembly and secretion; and gp3, gp6, gp7, gp8, and gp9 comprise the phage structural major and minor coat proteins. The intergenic region between genes IV and II does not code for proteins, but it contains signals for the initiation of synthesis of viral and complementary DNA strands and for phage assembly, termination of RNA synthesis, and the expression of the following genes II, X, V, VII, IX, and VIII. Furthermore, this region has been used as the site for the insertion of foreign DNA into the M13mp vectors. A smaller intergenic region between genes VIII and III also contains a transcription termination signal and a promoter for the expression of the following genes III, VI, I, XI, and IV. An excellent and exhaustive review on the genetic organization of M13 and related filamentous phages was written by Model and Russel in 1988 (13) (see also Suggestions for Further Reading).
Figure 1. The genome of wild-type bacteriophage M13. The approximate locations of the genes are indicated by Roman numerals.
3. Reproductive Cycle
The reproductive life cycle of bacteriophage M13 (depicted schematically in Fig. 2) starts with the adsorption of the phage to the tip of the F pilus of the host Escherichia coli, specifically mediated by gp3, the viral adsorption protein (14-16). The phage is subsequently transported to the cell surface, probably as a consequence of pilus retraction. Some host-encoded outer-membrane proteins (TolQ, TolR, TolA) are reported to mediate uncoating and DNA penetration (17, 18), probably at adhesion zones between the inner and outer membranes. Also, the viral gp3 is thought to play a role in facilitating the entry of the phage genome into the cell by the formation of a pore, enabling the entire host cell envelope to pass through (19). During the infectious entry of the phage, the hydrophobic gp6, which probably functions as a sort of sealing agent to preserve phage stability (5), is lost from the phage, resulting in a destabilized nucleoprotein particle ready for further disassembly. The major coat protein, and probably also the minor coat proteins, are stripped off from the phage and subsequently deposited in the inner membrane (20). During this process, the phage genome is injected into the cell cytoplasm.
Figure 2. Schematic illustration of some features of the reproductive life cycle of bacteriophage M13.
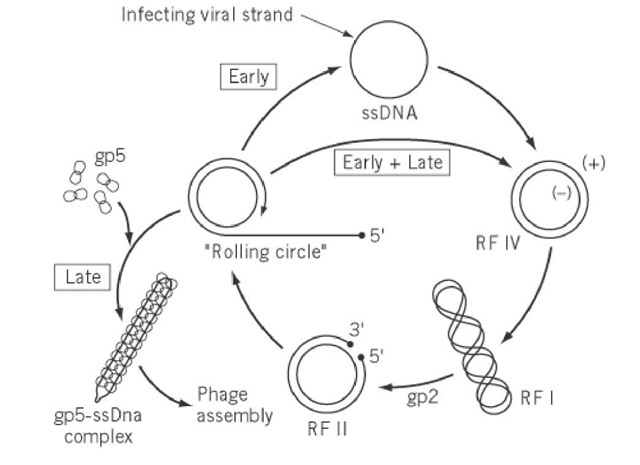
Following entry into the cytoplasm, the single-stranded phage genome is converted into a double-stranded supercoiled covalently closed replicative form (RF) DNA molecule by the action of host enzymes. This parental RF is the template for subsequent phage DNA replication by a rolling-circle mechanism (21), as well as for the transcription and translation of the 11 phage-encoded proteins (Fig. 3). At an early stage of infection, a pool of progeny RF molecules is generated under control of phage-encoded gp2. At a later stage, gp2, gp5, and gp10 regulate the levels of phage RF DNA and single-stranded DNA in the cell, which determines the rate of phage protein biosynthesis and the rate of phage assembly and concomitant extrusion, respectively. Upon binding of gp5, the newly synthesized viral strands are separated from the DNA replication machinery. The resulting single-stranded viral genome is encapsidated by the gp5 homodimers (22), but leaving the double-stranded packaging signal exposed, and is then ready to be assembled into progeny phage particles at the membrane-bound assembly site.
Figure 3. Schematic representation of the DNA replication cycle of bacteriophage M13. The early stage is characterized by the predominant synthesis of the replicative form (RF). Later in the infection cycle, the viral strand is withdrawn from the DNA replication machinery by gp5 and directed toward the phage assembly site.
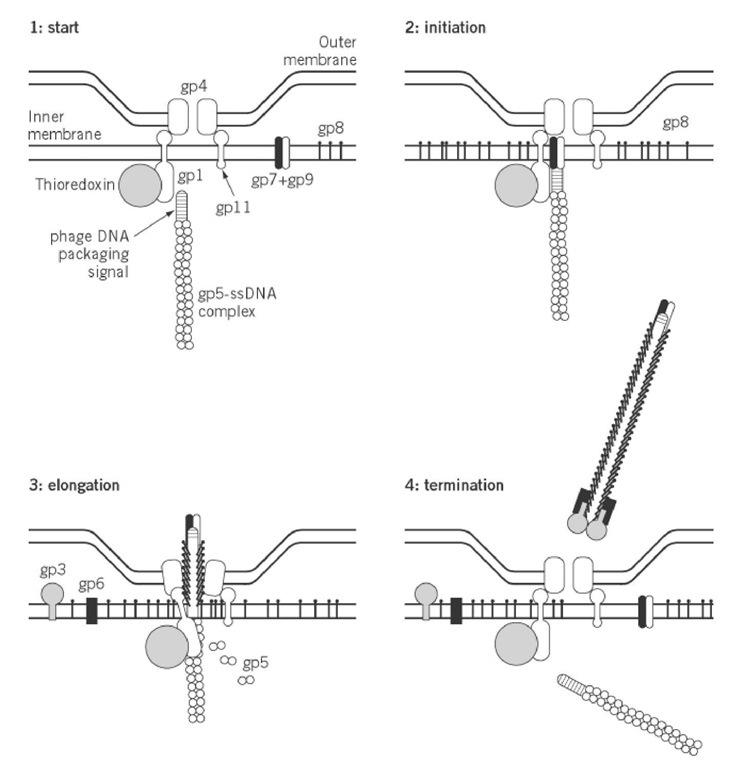
In contrast to most other bacteriophages, which are assembled intracellularly and are extruded by cell lysis, the morphogenesis of filamentous bacteriophage is concomitant with extrusion and without lysis of the host cell. Infected cells continue to grow and divide while extruding progeny phage particles into the medium. Filamentous phage emerge the infected cells at the assembly sites, which resemble adhesion zones between the inner and outer membranes (23). Three morphogenetic proteins gp1, gp4, and gp11 (the latter being an in-frame translated C-terminal part of the gp1), as well as at least one host-encoded protein (thioredoxin), are required for phage assembly. The proteins gp1 and gp11 are both associated with the inner membrane, and mixed multimers of them probably constitute the part of the extrusion channel that passes through the inner membrane (24, 25). A homomultimer of gp4 is thought to constitute the part of the extrusion channel passing through the outer membrane (26). As depicted in Fig. 4 these proteins interact and form a complex that could span the entire cell envelope.
Figure 4. Schematic illustration of the different stages of the membrane-associated assembly process of bacteriophage M13.
On initiating phage assembly, all the constituents come together around the inner membrane-associated gp1, which seems to be the "spider in the web" (see Fig. 4). All newly synthesized coat proteins are inserted into the inner membrane, where they are stored prior to being used in the assembly process (27). The exposed double-stranded DNA packaging (or morphogenetic) signal on 1 gp5-ssDNA complex interacts with gp1, which in turn triggers the formation and opening of the extrusion channel. Phage assembly might also be initiated by binding of gp7 and gp9 to the exposed double-stranded DNA packaging signal, probably mediated by gp1 (28). Furthermore, the associatio of thioredoxin with gp1 is a prerequisite for the elongation of the nascent phage. However, the main process during assembly of the new phage particle is the replacement of each gp5 monomer in the gp5-ssDNA complex by about two gp8 major coat-protein molecules, which protects the phage genome outside the cell. For this purpose, the initially membrane-bound gp8 should specifically recognize gp1 or gp11 in the assembly site.
Upon separating from the membrane lipids, and undergoing a conformational change from an L- shaped membrane-bound state to an aligned helical molecule, the gp8 molecules fit together with the other gp8 molecules, which all wrap around the phage DNA, thereby forming the coat of the new assembling phage. Meanwhile, the already assembled part of the new phage is extruded. The final, terminating step in assembly occurs by addition of gp3 and gp6 to close the phage filament, which is then released into the medium. Next, a voyage to another host cell can start.
4. Current State of Research, Future Prospects, and Implications of Interest
The information gained about the relatively simple, and therefore extensively characterized, filamentous M13 phage structure and reproductive cycle is very important for biology, because it can be translated to other more complex biological systems; the physical and chemical principles of interactions in biological processes will be the same. On the other hand, the versatile M13 phage is tl system of choice to study fundamental macromolecular interactions, because many kinds of differen processes are involved in filamentous phage infection, and ample detailed background information is already available. The study of the structure-function relationships of the various phage-encoded proteins, as well as the many mutual interactions in the formation of macromolecular assemblies, is therefore an attractive and promising field of research.
Employing state-of-the-art techniques in the fields of genetics, molecular biology, biochemistry, and biophysics, M13 phage and the related filamentous phages have had an enormous impact on the current research covering molecular recognition (4, 16), virus structure (6, 29, 30), protein-protein interactions (31-33), protein insertion across membranes (34, 35), DNA-binding proteins (36, 37), membrane protein assembly (38-41), and protein secretion (42, 43).