In classical genetics, the word locus referred to a position on a chromosome marked by a gene difference that has a phenotypic effect. Neither the locus nor the gene were thought to be subdivisible by recombination, and so locus and gene were treated almost as synonyms. The idea of complex loci arose from examples, especially from Drosophila and maize, of different mutations, apparently at the same chromosome locus, that give a range of different phenotypic effects, suggesting that the locus harbors some rather complex kind of gene.
With our modern knowledge, the word locus can take either of two very different meanings. It may mean just the genetically mapped site of a mutation, which, at the molecular level, is quite likely to be no more than a single base-pair change within the long DNA sequence of a gene. But, in the context of complex loci, it can mean a relatively short chromosomal segment (not frequently split up by genetic recombination) that contains a cluster of genes among which there is some functional connection. So complex loci, though of somewhat confused etymology, is a useful heading under which to review the evidence for chromosomal units of function at a level higher than that of the individual gene.
This article does not cover examples of loci that mutate to give various phenotype effects when the varied effects are due to mutations in a single gene that has multiple functions. Such genes that encode "multiheaded" enzymes (see Multifunctional Proteins) are sometimes called "cluster genes" in fungal genetics to distinguish them from the gene clusters with which they may initially be confused. Some examples are reviewed under Allelic Complementation. Nor are we concerned with single genes that have multiple modes of intron splicing, which yield alternative messenger RNAs (see examples under Gene Structure). And we obviously cannot include groups of functionally diverse genes that just happen to be very closely linked, for that would make virtually all genes parts of one complex locus or another. To qualify as a complex locus, in our present sense, a gene cluster must have some kind of unity in structure, in function, or in both.
Gene clusters that fulfil this criterion have been found in fungi, flowering plants, Drosophila and mammals, and this article reviews examples from each of these kinds of organisms. We do not deal here with bacteria. Operons, which might be viewed as complex loci but are not usually so termed, are dealt with under their own heading. Also excluded from this article are highly repetitious tandemly arrayed genes required by the cell in very high dosage. The preeminent example is ribosomal RNA genes in the nucleolus organizer chromosomal regions of eukaryotes. Histone genes are also highly repetitive and are concentrated in clusters in some organisms, including Saccharomyces yeast. This is arguably more repetition than complexity.
1. Fungi
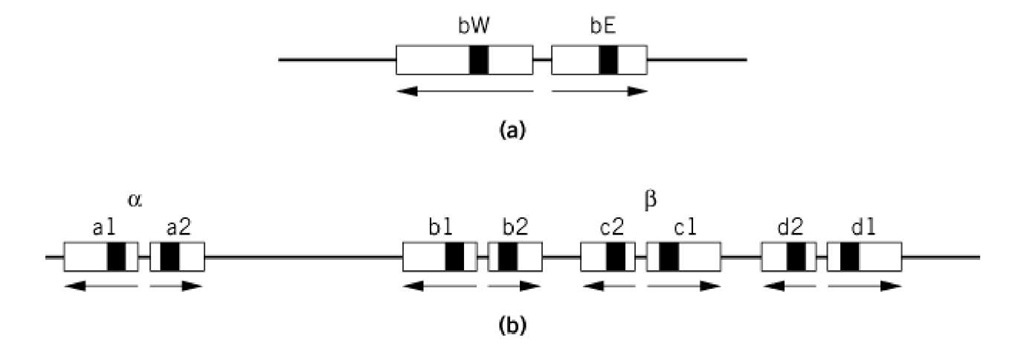
In fungi, the self-incompatibility and cross-compatibility in sexual mating in many species is determined by mating-type loci, which virtually always consist of more than one gene. A relatively simple example is the maize smut fungus, Ustilago maydis, whose two loci, a and b, control the specificity of sexual fusion and the formation of an invasive dikaryotic mycelium after fusion, respectively. The a locus has two "alleles," al and a2, each of which consists of two genes, one that encodes a polypeptide pheromone and the other a pheromone receptor. The al pheromone is recognized by the a2 receptor and the a2 pheromone by the a1 receptor. There is no close homology between al and a2, and they do not recombine with each other. The b locus exists in numerous alternative forms. Each consists of two closely spaced, divergently transcribed genes called bE and bW that encode the two components of a dimeric transcriptional activator. But dimerization occurs only between bE and bW products of different mating types (1).
The ink-cap mushroom fungus, Coprinus cinereus (like Ustilago, a member of the Basidiomycete class), also has two mating-type loci, A and B. The A locus, which controls the ability to form a fruiting dikaryon, is the most thoroughly investigated. Here the situation resembles that at the Ustilago b locus, except that genes that encode dimerizing monomers, thought to be transcriptional activators on the basis of their homeobox-like sequence motifs, are distributed in multiple copies between two subloci, Aa and Ab, between which rare recombination occurs (2) (Fig. 1). Again, dimerization occurs only between the products of different mating types.
Figure 1. The structures of mating type loci in Basidiomycete fungi. (a) The b locus of Ustilago maydis. The bWand bE gene products form W-E protein dimers to activate transcription of genes necessary for sexual development. Each exists in multiple allelic forms. Effective dimers are not formed by W-E combinations from the same strain. Gene segments that encode homeodomain-like protein sequences are filled. (b) The archetypal A mating type locus of Coprinus cinereus. The system works in principle as in the Ustilago b locus, except that here there are four pairs of genes, a, b, c, and d, any one of which can function in promoting fertility. The archetype is a composite of known mating-types, all of which are missing one or another part of it.
Ascomycete fungi, such as Neurospora crassa and Saccharomyces cerevisiae, simpler than Basidiomycetes, have only two mating types (A and a in Neurospora, a and a in Saccharomyces), but the genetic loci that determine them are still complex. Neurospora A contains three genes (Fig. 2) and Saccharomyces a contains two, though each of the a mating-type loci has only a single gene. The two alternative mating-type loci in both organisms are completely nonhomologous, and the same is true for the Ustilago a locus. It has been customary to call the alternative occupants of these mating-type loci "alleles," but in the light of present knowledge this terminology is inappropriate because it implies different forms of a single gene. The new word idiomorphic has been suggested for the relationship between quite different genes or gene clusters that are alternative occupants of the same locus (3). Much the same applies to the yeast mating-type system that has the additional complication of mating-type switching.
Figure 2. The mating type loci of the Ascomycete fungus Neurospora crassa. A1 and al, quite unrelated in sequence, act in a complementary way to permit sexual reproduction. A2 and A3 function in postfertilization fruiting-body development.
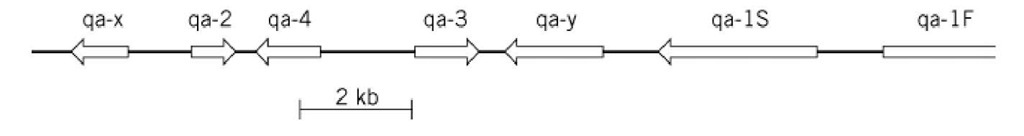
Fungi also provide examples of another kind of locus complexity, the tight clustering of separate genes that co-operate in a particular area of metabolism. The Neurospora crassa Qa cluster is a particularly good example (Fig. 3). Here a DNA segment of only 18 kbp harbors seven different genes, the transcription of which is increased 50- to 1000-fold when the fungus is forced to grow using quinate as a sole carbon source. Three of these genes encode the three enzymes that, acting in sequence, oxidize quinic acid to protocatechuic acid. Two genes encode regulatory proteins that control the transcription of the whole cluster. One gene is a transcriptional activator and the other a repressor that cancels the function of the activator but whose own effect is nullified by binding to quinate. The remaining two genes have no identified function and are recognized solely as open reading frames (4). The significance of the gene clustering is not fully understood, but it seems plausible to suggest that derepression of transcription is facilitated by an induced unfolding of nucleosomal structure that extends across the entire complex.
Figure 3. The Qa (quinate-utilization) gene cluster of Neurospora crassa. The genes qa-3, qa-2, and qa-4, respectively, encode quinate dehydrogenase, catabolic dehydroquinase, and dehydroshikimate dehydratase, which act sequentially to degrade quinate. qa-1S and qa-1F encode regulatory proteins, that respectively repress and activate transcription of the whole cluster. qa-Y, it is thought, encodes a quinate transporter protein, and qa-Xhas no known function (4).
2. Flowering plants
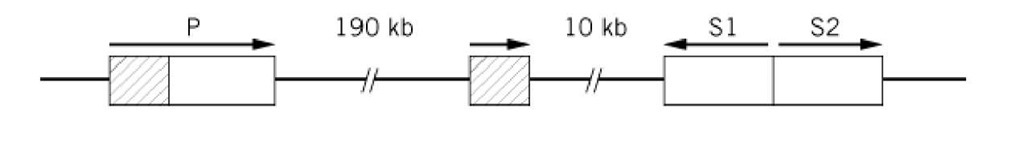
A long-standing example from what used to be, pre-Arabidopsis, the geneticists’ favorite plant, is the R locus of Zea mays that is necessary for pigmentation and encodes transcriptional activators for genes that encode pigment-synthesizing enzymes. It was an early example of locus complexity because it has two independent functions. Some recessive loss-of-pigment alleles affect only the red color in the plant whereas others affected only the seed. Analysis at the DNA level has revealed the situation shown in Fig. 4 (5). The R locus contains three genes. One gene (P) is specific for plant pigment, and the other two (S1, S2) apparently duplicate the function in seeds. Presumably their distinct tissue specificities are due to different promoter/enhancer sequences. There is also a partial and presumably functionless duplication of P. An origin by duplication and subsequent partial divergence of function is the most obvious explanation for the clustering of these functionally related genes, although the opposite orientations of S1 and S2 are a puzzle.
Figure 4. The complex R locus of Zea mays. P controls red pigment synthesis in the plant generally and the duplicate S genes in the seed. S1 and S2, transcribed divergently, are each similar to P except that they lack the segment shown hatched. This segment is present as a tandemly duplicated fragment, presumably nonfunctional. Sequence information suggests that this scrambled arrangement is due to the activity of a transposable element (5). "Alleles" of R, Rg (red seed, green plant), and rr (colorless seed, red plant) are, respectively, due to loss of P and S function. The P and S components can be separated by meiotic crossing over at frequencies on the order of 10-4 to 10-3.
A long-standing problem in plant biology has been the various systems of self-incompatibility, often controlled by what are multiple alleles at a single locus, usually called S. The general principle is that pollination is prohibited if the pollen grain and stigma (or, in the Brassica family, the pollen mother cell and stigma) have an allele in common. The Brassica S locus, one of the best analyzed, contains two genes, each present in the population in multiple allelic forms, that control the incompatibility response of the stigma. A third S locus gene, expressed in the pollen and determining pollen specifity, has also been identified (6). Here, as in the analogous self-incompatibility in fungi, the functional reason for the multiple genes all in the same locus is obvious. For the system to work, the different specificities have to be mutually exclusive in the haploid genome.
3. Drosophila
In the fruit fly, Drosophila melanogaster, a classical example of locus complexity, perhaps originally generated by tandem duplications, is provided by the scute (sc) and achaete (ac) families of mutations (7, 8). These affect the bristles in the wings and mid-thorax of the fly, really parts of the fly’s neural system, the origin of which can be traced back to the transcription of scute and achaete, usually both together but sometimes one more than the other, in rather precisely positioned bristle mother cells in the wing imaginal disc of the larva. It seems that sc and ac largely duplicate each other’s functions and, before molecular analysis, the basis for recognizing two different genes was not clear. Different mutations in the sc/ac locus eliminate different bristles or different combinations of bristles, and, to the extent that their effects are nonoverlapping, complement one another in trans. Now that the sc/ac locus has been analyzed at the DNA level, it is seen that there are six genes within the approximately 100 kbp that comprise the sc/ac complex (7, 8) (Fig. 5), of which at least four are involved with the neural/bristle system. sc and ac apparently have the major roles. Most of the mutations that affect the positioning of bristles actually fall between the genes. Several are chromosomal rearrangements that have breakpoints within the ac/sc complex, and it is thought they exert their effects by separating ac and/or sc from enhancer sequences, several of which have been identified in the sc/ac region. The patterns of transcriptional activation of sc and ac and hence the positioning of bristles, it is thought, reflect the distribution of a number of enhancer-binding proteins, the identity of which remain to be established. The genes and the enhancers together may be seen as an integrated system for responding to a pattern of positional information set by the products of other genes.
4. Mammals
Figure 5. The scute-achaete (sc/ac) complex at the tip of the Drosophila melanogaster X-chromosome. Genes that have functions in neural/bristle development are shown as filled arrows, and other genes, apparently unrelated in function, as o arrows (y = yellow). Thin vertical arrows indicate the breakpoints of segmental rearrangements that affect the bristle phe (sc6 is a deletion). Regions that include enhancer sequences are shown hatched. The different bristle phenotypes are belie be due to separation of sc and/or ac from enhancers.
Many clusters of related genes have originated through tandem duplication and subsequent functional divergence. Obvious examples are the globin gene clusters in animals and humans (see under Globins, Gene Structure). The b-globin gene cluster has a common ancestral origin and is also functionally integrated through the locus control region.
The clustered HOX genes, which are of profound importance in animal development and encode homeobox-containing proteins, were discovered in mammals and now in other animal groups through their significant similarities to the genes of the linked bithorax and Antennapedia complexes of Drosophila. Five unlinked clusters of HOX genes in mouse and humans each consist of up to 13 tandemly arranged genes within a region on the order of 100 kbp of DNA. Through their homeobox-encoding segments, the genes of one cluster have only a very limited degree of sequence similarity to each other but have a greater resemblance to genes in corresponding positions in other clusters in the same species or even in different species. Because they were not identified through the classical genetic route of mutation mapping, the HOX clusters of mammals are not usually called loci though, confined as they are within chromosomal segments that correspond to a small fraction of a centimorgan, they are sufficiently tightly linked to qualify for that title, and they also fulfil the criterion of functional integration. The almost uncanny correspondence between the linear sequence of genes within each cluster and the anterior-posterior order of the body segments that the genes affect clearly indicates some function for the clustering and ordering of the genes, though this is still mysterious.
The mammalian major histocompatibility complex (MHC), intensively investigated in mouse, and its human equivalent the human leucocyte antigen (HLA) complex, is encoded by genes spread over rather too large a section of chromosome to be called a locus. Indeed, the positions of individual antigen-encoding genes within it are themselves often called loci. In humans it encompasses about four megabases of DNA and 3 to 4 centimorgans. But it may be regarded as at the upper end of the size range of clusters of functionally related genes, and so it is relevant to mention it here. A diagram showing of the structure of the complex, and its scale in relative to the whole chromosome is shown in Fig. 6. Because there is significant homology among the different genes, at least within each of the two main subclusters, the close linkage is likely to be due to duplication by short-range transposition and subsequent diversification. But it may also have been maintained by natural selection, because their continued coupling is advantageous if certain combinations of alleles of different genes confer disease resistance (10).
An entirely different kind of complex locus is involved in determining hypervariable proteins of the mammalian immune system, immunoglobulins and T-cell receptor proteins. These are not really gene clusters but rather banks of gene components that are assembled by DNA splicing reactions in a large number of different ways to provide coding sequences for proteins of almost any required specificity. In mammals, as exemplified by mouse, there are six known rearranging complexes of the mouse immune system, three (heavy-chain and two light-chain) for immunoglobulin components and three (a, b and g polypeptides) for T-cell receptor components. A diagram to scale of the completely sequenced human b-chain locus that extends over 600 kb is shown in Fig. 7 (11). Here the functional reason for clustering is obvious. It is only surprising that the sequences destined to be spliced together are as widely separated as they are. Presumably they are brought close during the splicing process by the formation of some very extensive DNA loops.
Figure 6. The arrangement and spacing of genes within the major histocompatibility complex (MHC) of mouse. Class I genes encode transplantation antigens mediating graft rejection (K, D, L), or other cell-surface antigens (Qa, Ti). Class II genes encode antigens involved in the immune response. Class III genes encode components of the complement system. Meiotic recombination within the complex is infrequent but not very rare (genetic map distances are shown in centimorgans, cM).
Figure 7. The arrangement and spacing of the part-genes which, when appropriately spliced together, encode b-chains of the human T-cell receptor. Splicing of DNA during T-cell differentiation brings together one C "constant", one J ("junction"), one D ("diversity"), and one V ("variable") sequence to encode a large number of alternative b-chains. J on the map stands in each case for six or seven short alternatively used junction sequences. The other marked elements are all single units. All are transcribed/translated from left to right, except for the rightmost V which is inverted and doubtfully functional. The splicing occurs in stages, first D to J, then DJ to V, and finally VDJ to C. The diagram has been simplified by omitting the pseudogenes and incomplete V sequences, which approximately equal the functional genes in number and are interspersed among them. T = tripsinogen gene, the presence of which, in multiple copies, within the b-chain complex, has uncertain functional significance.