The pentose sugar L-arabinose is distributed widely as a component of plant polysaccharides in pectins, gums, and cell walls. It is also present in smaller amounts in the cell walls of some bacteria. Thus it is likely that, in its natural environment in the vertebrate intestine, the bacterium Escherichia coli periodically encounters arabinose released through degradation of these polymers (1). Not surprisingly, E. coli, like many other bacteria and fungi, has ara genes that encode the enzymes and transport proteins necessary to utilize L-arabinose as a source of carbon and energy.
The ara genes of E. coli have been of exceptional importance to biology for several reasons. If the lac operon is the exemplar of regulation of gene expression by repression of expression, the ara operon is the counterpart as the model for positive control of gene expression, even though more complete analysis has revealed that each has both positive and negative features. (An operon is one gene or a linear sequence of related genes that are transcribed as one unit from a common initiation point [see Transcription]. The transcript is then translated into protein products corresponding to the separate genes.) The first clear example of DNA looping as a feature of transcription control was the ara operon. The hypothesis of positive control of the ara operon was a challenge to establishment scientists, and its validation was the end of a fascinating story in the history of science. Moreover, the details of the operon are complex and interesting in themselves.
1. ara Operon of Escherichia coli
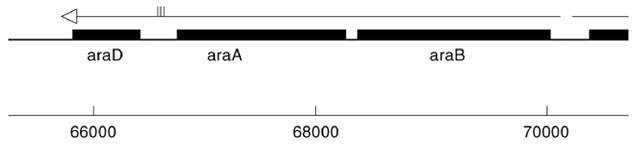
The classical ara operon of the bacterium Escherichia coli comprises three genes, araBAD (Fig. 1). This and four other operons, araC, araE, araFGH, and araJ, are uniquely associated with metabolism of L-arabinose (Table 1). Each set of genes is under control of the activator protein AraC, the product of the araC gene. The function of each gene is known, except for araJ, which may encode a protein that processes or transports an arabinose-containing polymer, or pumps potentially toxic arabinosides from the cell.
Figure 1. The positions of the araBAD operon and the araC regulatory gene on the E. coli chromosome. Transcription o nucleotide-pair region between araB and araC. Nucleotide location on the E. coli chromosome is shown beneath the gen the three inverted repeat REP sequence pairs that are assumed to produce three self-paired hairpin structures in the mRN entire chromosome contains approximately 4,639,221 nucleotide pairs.
Table 1. ara Genes and Gene Products
|
Gene |
Location on the E. coli chromosome (minutes)2 |
Size of gene (amino acid codons) |
Activity of product |
|
araA |
1.4 |
500 |
L-arabinose isomerase |
|
araB |
1.5 |
566 |
L-ribulokinase |
|
araC |
1.5 |
292 |
AraC regulatory |
|
protein |
|||
|
araD |
1.4 |
231 |
L-ribulose-5-phosphate-4-epimerase |
|
araE |
64.2 |
472 |
L-arabinose transport, |
|
low affinity |
|||
|
araF |
42.8 |
329 |
L-arabinose transport, high affinity |
|
araG |
42.7 |
504 |
L-arabinose transport, |
|
high affinity |
|||
|
araH |
42.6 |
329 |
L-arabinose transport, high affinity |
|
araJ |
8.5 |
394 |
Transport or processing of polymer? Efflux of toxic arabinosides? |
a Location is based on a 100 minute circular chromosome.
1.1. Uptake and Utilization of Arabinose
Two independent systems deliver arabinose from the environment across the cell membrane into the cell. araE encodes a membrane protein that mediates arabinose uptake via proton symport and is the lower affinity transporter (2). The araFGH operon encodes a periplasmic arabinose- binding protein (araF), a probable ATPase subunit (araG), and a membrane protein (araH), which together mediateATP-driven arabinose transport. This transporter shows higher affinity for arabinose
(2). The araFGH operon encodes a periplasmic arabinose- binding protein (araF), a probable ATPase subunit (araG), and a membrane protein (araH), which together mediateATP-driven arabinose transport. This transporter shows higher affinity for arabinose![]() than AraE, but lower capacity (2, 3).
than AraE, but lower capacity (2, 3).
Internal arabinose is converted in three steps to D-xylulose-5-phosphate, a metabolyte in the pentose-phosphate shunt pathway and one that is not unique to arabinose metabolism. The enzyme mediating the first step, L-arabinose isomerase, has a low affinity for arabinose with a KM (Michaelis constant) of 60 mM (4); this suggests that cells growing on arabinose have a very high internal arabinose concentration (5). The glucose-specific phosphotransferase enzyme IIAGlc when unphosphorylated inhibits the isomerase (unpublished results cited in 6) This inhibition may be one of the causes of the preferential use of glucose when both arabinose and glucose are in the environment.
The product of isomerase activity, L-ribulose, is converted to L-ribulose-5-phosphate by L-ribulokinase, and the phosphorylated compound is converted in turn to xylulose phosphate by the epimerase encoded by araD. Arabinose inhibits growth of araD mutants on other nutrients, presumably because accumulation of a high concentration of ribulose phosphate is toxic. Thus secondary mutants lacking isomerase or kinase activity as the result of araA, araB, or araC mutation, and therefore not forming ribulose phosphate, can be selected by plating an araD population on broth plates containing arabinose (7).
1.2. Regulation of ara Operon Expression
In the absence of arabinose, the ara genes are essentially not expressed, except for the regulatory gene, araC. On exposure to L-arabinose, all of the ara genes are activated, transcription of araBAD begins within five seconds, and the Ara proteins appear within several minutes, allowing growth on the sugar (5). The true inducer has been shown by X-ray crystallography of the inducer-AraC protein complex to be -L-arabinose (8). L-lyxose also induces the ara genes (9). However induction of the ara genes by lyxose is unlikely to occur in nature because L-lyxose is rare. araC mutants have been obtained that are inducible by D-fucose or by -methyl-L-arabinoside, normally inhibitors of induction (1, 10, 11).
In vivo studies using cells lacking both arabinose transport systems showed the KM for induction by arabinose to be unexpectedly high, nearly 10 mM (11), although, as noted above, the internal arabinose concentration is likely to be higher than this during growth on arabinose because of the low affinity of the isomerase for the pentose. The high concentration-dependence of induction suggests that there might be natural selection against a mutant isomerase with a better affinity for arabinose.
Complementation studies showed that the presence of AraC protein is necessary for araBAD gene expression (12, 13), a discovery unanticipated by those who believed all gene activation resulted from removal of repressors. Later, AraC was shown to be required for expression of the other ara operons as well (except for araC). araC is the prototype for a large family of regulatory genes that share sequence similarity in the DNA binding region and are homologous and presumably related through evolution (14).
Cyclic AMP (30,50-cyclic AMP, cAMP) bound to cyclic AMP receptor protein (CRP) is also a positive regulator for all the ara operons. Expression of many genes is controlled by availability of CRP-cAMP; lack of expression due to low CRP-cAMP is referred to as catabolite repression. In vitro transcription of araBAD mimics that in vivo in that it requires both the AraC and CRP regulatory proteins with their bound ligands. Analysis of transcription in vitro, and further in vivo studies, have given a broad understanding at the molecular level of control of araBAD transcription although details remain to be determined (5).
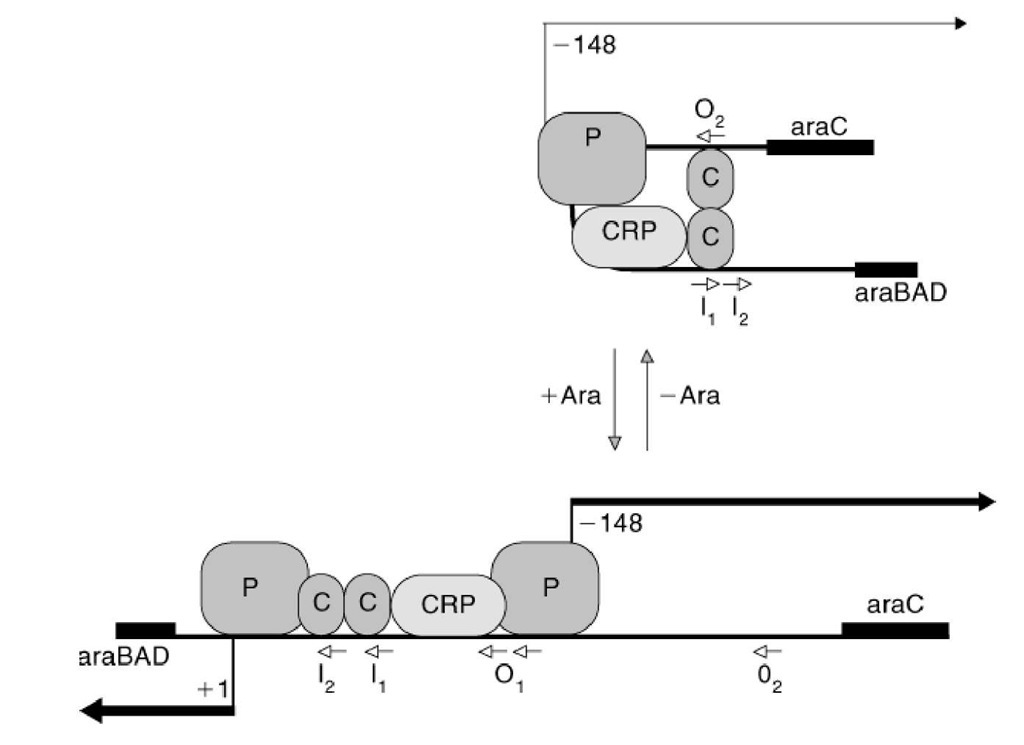
The AraC protein structure has two domains connected by a flexible polypeptide linker. The N-terminal domain binds arabinose and is responsible for formation of the active dimeric form of the protein. The C-terminal domain binds DNA at specific sites, with similar sequences, upstream of each ara operon. In the regulatory region between the divergently transcribed araBAD and araC operons (Fig. 1), there are five sites at which AraC can bind (Fig. 2).
Figure 2. Activation of transcription of the araBAD operon by arabinose. CRP refers to CRP-cAMP, and P refers to RNA polymerase. AraC protein is shown as a dimer bound to I1and O2in the absence of arabinose and to I1and I2in the presence of arabinose. I1, I2, O1 (a double site), and O2are all potential binding sites for the AraC protein and conform to the consensus sequence 50;-TAGCN7TCCATA-30 (reading in the direction of the arrows) (16) although there is considerable variation among sites. Note how the mode of pairing of the dimers differs depending on the presence of arabinose (5, 8). On addition of arabinose, transcription of araBAD is initiated and expression of araC is increased (bottom). After a few minutes AraC-arabinose dimers are thought to bind to O1 and O2 so as to form a new DNA loop and reduce araC transcription to that characteristic of cells in the absence of arabinose (not shown).
In the absence of arabinose, the two DNA-binding domains of the AraC dimer are oriented so that they can not readily bind both of the adjacent I sites at the same time. Instead, the AraC dimer contacts I! and O2, thereby forming a DNA loop within the region between araB and araC (Fig. 2) (15). On addition of arabinose, the dimer undergoes a conformational shift such that the two DNA-binding domains preferentially bind ^Ij (Fig. 2) (16). The presence of AraC at I2 stimulates addition of RNA polymerase and open complex formation, and transcription of araBAD commences, if CRP-cAMP is present (5). Although this model was proposed and refined before detailed structural information was available, X-ray crystallographic studies of the AraC ^-terminal domain and linker support the model (8).
Addition of arabinose also affects expression of araC. When the I1-O2 loop is opened, transcription of araC is accelerated (Fig. 2). After a few minutes, AraC-arabinose dimers are thought to reform DNA loops, this time by bridging O1 and O2 (17, 18). O1-O2 looping does not regulate araBAD transcription, but interferes with RNA polymerase binding and initiation at the araC promoter; this reduces the rate of araC transcription to that characteristic of cells in the absence of arabinose (5). araC is controlled by CRP-cAMP as well. CRP-cAMP binding increases, but is not essential for, araC transcription; it is necessary for substantial expression of the other ara operons.
The mechanism by which AraC-arabinose, CRP-cAMP, and RNA polymerase interact to trigger transcription is not clear. Bound CRP-cAMP helps to open the I^ repression loop on addition of arabinose, but it probably activates RNA polymerase binding or initiation as well, either by direct contact or indirectly through contact with AraC (5). (CRP-cAMP does not aid AraC binding.)
Several kinds of CRP-cAMP-independent mutants have been obtained. One class (rpoD) has an altered RNA polymerase sigma factor-70 subunit (19), while another (araC) is altered in AraC itself (20). Other mutants (araI) are transcribed by RNA polymerase independently of the AraC or CRP proteins, and they result from changes at the RNA polymerase binding site in the araBAD promoter (21).
2. Other Mechanisms for Arabinose Degradation
The pathway for arabinose degradation that is common to E. coli and many other bacteria is not the sole means for utilizing L-arabinose in the biological world, nor is the regulatory model described above the only means for control of ara enzyme synthesis. At least five different pathways exist (1), and different regulatory systems are found even among organisms using the same pathway. Although the work on other organisms is still at an early stage, it is known that the gram-positive bacterium, Bacillus subtilus, utilizes the same degradatory pathway for arabinose utilization as E. coli but appears to use a simple repressor as the means of control of ara gene expression (22). The fungus Aspergillus uses a completely different pathway for arabinose utilization and, not surprisingly, appears to have different control systems as well (23). It will be remarkable if the ara genes in these other organisms are regulated by the simple repressors once hypothesized for the ara genes in E. coli.