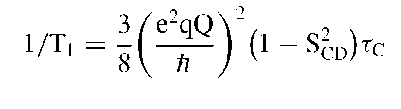Biological membrane A very thin sheath of biological material (thickness ~10 nm to 15 nm) which constitutes the envelope of living cells and also of intracellular organelles, separating them from the environment. Membranes are made up from a lipid bilayer into which proteins are embedded. They are highly organized but are nevertheless fluid enough to allow considerable translational, rotational, and flexing movements of the constituent lipid and protein molecules.
Lipid bilayer A double layer of lipid molecules organized in a tail-to-tail arrangement. It is an anisotropi-cally ordered fluid that has a number of properties in common with smectic liquid crystals. The normal to the surface of the lipid bilayer constitutes an axis of motional averaging. From an optical point of view, bilayer membranes thus behave like uniaxial crystals with the bilayer normal as the optical axis (director axis n). All fast molecular motions such as rotational and flexing movements are thus characterized, on the average, by a cylindrical symmetry with the bilayer normal as the axis of motional averaging.
Deuterium nuclear magnetic resonance (2H-NMR) By means of chemical synthesis or biochemical incorporation, protons in lipid molecules can be selectively replaced by deuterium atoms. Since the van der Waals radii of the two isotopes are identical, this substitution leaves the membrane virtually unchanged, which is in contrast to other bulkier reporter groups. Deuterium nuclear magnetic resonance provides information on the order and mobility of the molecule. Structural information comes from the deuterium quadrupole splitting, dynamic information is derived from NMR relaxation times.
Quadrupolar splitting (Avq) The deuterium nucleus has a spin = 1 and, due to its electric quadrupole moment, the anisotropic motion within the membrane will give rise to a quadrupole splitting, Avq (kHz). In an un-oriented sample, as most membrane preparations are, the deuterium quadrupole interactions give rise to a characteristic powder pattern. The spectrum has two distinct peaks, the separation of which is the so-called deuterium quadrupole splitting, AvQpowder. The deuterium quadrupole splitting may by used to calculate the deuterium order parameter SCD according to
The static deuterium quadrupole coupling constant is (e2qQ/h) = 170 kHz for aliphatic carbon-deuterium (C—D) bonds. A change in the residual quadrupole splitting can be caused by two different mechanisms. First, the angle of the molecular fluctuations may increase or decrease, secondly, the molecule may undergo a conformational change which alters the orientation of the C—D bond vector with respect to the bilayer normal.
Order parameter (Scd) The deuterium order parameter is a measure of the motional anisotropy of the particular C—D bond investigated and yields its time-averaged orientation. If © denotes the instantaneous angle between the C—D bond and the direction of the bilayer normal then SCD is defined as
where the bar denotes a time average. Order parameter (Smoi) Assuming an axial symmetry of the segment motion SCD can further be related to the molecular order parameter Smol according to
If the chains are fixed in an all-trans conformation and are just rotating around the long molecular axis, the molecular order parameter would be unity. The other extreme is that of a completely statistical movement through all angles of space, leading to Smol = 0. This simple statistical interpretation of SCD is not possible if specific geometric effects come into play as, for example, in the case of the cw-double bond.
Order profile of the lipid bilayer It shows the variation of the order parameter, Smol or SCD, with the position of the segment in the chain and is an expression of the average angular fluctuations around the bilayer normal.
Spin-lattice relaxation time (Ti) The spin lattice relaxation time depends on both the ordering (SCD) and the rate of motion (correlation time, rC). Assuming a motion sufficiently characterized by a single correlation time, tc, the following expression holds for the short correlation time limit:
Phosphorous nuclear magnetic resonance (31P-NMR) No isotope labeling is required for 31P-NMR spectroscopy. The chemical shielding anisotropy, Aa, in 31P-NMR is comparable to the deuterium quadrupole splitting in 2H-NMR and can be determined from the edges of the spectrum.