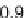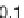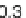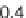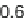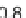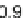Biology Reference
In-Depth Information
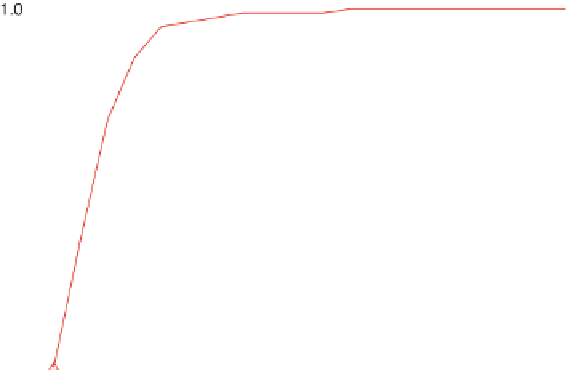
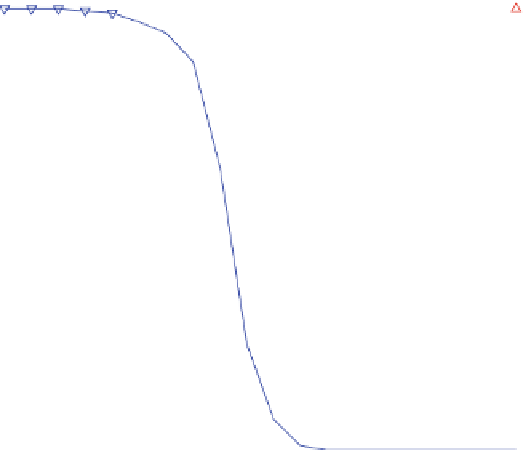
Fig. 3 Relative grammar-based distance classification thresholds at the species level. Given a known set of
16S Ribosomal RNA sequences in which multiple sequences belong to the same species, a binary classifica-
tion is performed for varying grammar-based complexity thresholds. The classification procedure is to
compare the pairwise distance between two sequences against a threshold. If the distance is less than the
threshold, the two sequences are classified as belonging to the same species. If the distance is greater than
the threshold, the two sequences are classified as belonging to different species
results of which are presented in Figs.
3
and
4
. Given a known set
of 16S Ribosomal RNA sequences in which multiple sequences
belong to the same species Fig.
3
or genus Fig.
4
, a binary classifi-
cation was performed for varying thresholds. The classification
procedure was to compare the pairwise distance between two
sequences against a threshold. If the distance was less than the
threshold, the two sequences were classified as belonging to the
same species/genus. If the distance was greater than the threshold,
the two sequences were classified as belonging to different species/
genera. So, a reasonable threshold might be more on the order of
0.30. Dissimilar sequences start exhibiting relative distance scores
above 0.45.
After the alignment order has been determined, progressive align-
ment is accomplished by performing repeated pairwise alignment.
Referring to the right half of Fig.
1
, the sequences being aligned
may be: (1) two sequences that belong to the same similarity group,
or (2) two consensus sequences that represent the agreed-upon
average sequence of two dissimilar groups. The following options
provide a means for adjusting the amount of the alignment matrix
3.6 Alignment
Heuristic Options