Biomedical Engineering Reference
In-Depth Information
are cross-linked to it. These are then immunoprecipitated with specific
antibodies, the cross-link reversed, and the proteins identified on con-
ventional SDS-PAGE gels by western blotting.
DNA footprinting
DNA footprinting is a method for identifying the sites on the DNA
that are recognized by DNA-binding proteins. There are a number of
different DNA footprinting methods that allow for the detection of DNA
binding proteins under a variety of conditions. One of the most common
footprinting methods is DNase I footprinting. The DNA from a restriction
fragment is labeled at one end with and is subjected to partial hy-
drolysis by DNase I in the presence and absence of the bound protein.
The fragments produced will be different at the site of binding because
the bound protein will protect the DNA from cleavage. The protected
site is visualized as a gap in the ladder, the footprint, which identifies the
region bound by the protein.
Chromatin immunoprecipitation (ChIP)
ChIP is a method used to examine whether a particular protein is
bound to a given DNA sequence in vivo. Unlike EMSAs, which deter-
mine whether a particular transcription factor CAN bind to a DNA se-
quence, ChIP assays can reliably tell an investigator IF a DNA sequence
is occupied by a protein of interest under physiological conditions. In this
method, DNA-binding proteins are cross-linked to DNA (using formalde-
hyde), the chromatin is isolated and sheared into fragments with their
accompanying bound proteins. Specific antibodies are then used to im-
munoprecipitate the bound proteins with their corresponding DNA se-
quences. The cross-linking is reversed and, to determine if the DNA of
interest has been immunoprecipitated, specific PCR primers are used
to amplify the DNA.
Biotinylated DNA pulldown assays
This assay was developed to help improve the ability to identify larger
protein complexes associated with particular DNA sequences. In this
assay, biotinylated DNA (DNA can be biotinylated on the ends of
each DNA strand) is incubated with nuclear extracts. The complex can
be removed from other proteins (“pulled down”) in the extracts using
streptavidin-agarose beads. The proteins can then be dissociated from
the complex using SDS-PAGE sample buffer and resolved on these gels.
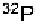

Search WWH ::

Custom Search