Biomedical Engineering Reference
In-Depth Information
shows an example of an EMSA using a probe that contains a binding
site, called which interacts with a family of transcription factors col-
lectively called The two shifted bands represent the binding of
two different complexes. To prove specificity of binding in EMSA
assays, competition experiments are performed. First, the binding of the
extracts can be performed in the presence of “cold” competitors (an un-
labeled identical probe added in excess), which demonstrates that the
shift is not nonspecific. Second, when a mutant probe with an identical
sequence except in the DNA binding site is used as a cold competitor,
binding is not inhibited.
To identify the DNA binding proteins in nuclear extracts that are bound
to the DNA probe, antibodies specific for known transcription factors can
be used. Two common methods employing specific antibodies for this
purpose are the supershift assay and UV cross-linking and immunopre-
cipitation.
Supershift assay
This is a variation of the EMSA in which antibodies are used to iden-
tify specific proteins associated with DNA probes used in EMSA. After
binding of nucleic acids to the radioactive probe, antibodies to specific
proteins are added to the complex. If the protein is present, the migration
of the complex will be altered. Specifically, the presence of the antibody
will further retard the migration of the complex, resulting in a band on
the gel that migrates more slowly than the corresponding band without
the antibody. As a result, the band appears shifted up, hence the name
“supershift”. Note, however, that some antibodies cannot be used for this
supershift assays since some can disrupt the binding of the protein to
the DNA.
Supershift assays are important not only to confirm the identity of the
protein interacting with a DNA sequence, but also are useful to iden-
tify proteins in complexes that do not necessarily interact with the DNA.
While active transcription complexes consist of proteins that interact di-
rectly with specific DNA templates, they often contain other proteins that
interact with these proteins (via protein-protein interactions). Examples
of these include coactivators. Supershift assays provide one way to de-
tect some of these additional proteins, although large complexes are not
frequently resolved by either standard EMSA or supershift assays.
UV-crosslinking and immunoprecipitation
In this variation of the EMSA, nuclear extracts are incubated with
the dsDNA template, and then the proteins that are bound to the DNA

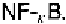

Search WWH ::

Custom Search