Biomedical Engineering Reference
In-Depth Information
$ head offset.txt
ENSMUSG00000090025:ENSMUST00000160944
$ head offset.txt
ENSMUSG00000090025:ENSMUST00000160944 0.007850
ENSMUSG00000090025:ENSMUST00000160944 0.007850
ENSMUSG00000090025:ENSMUST00000160944 0.021899
ENSMUSG00000090025:ENSMUST00000160944 0.021899
ENSMUSG00000090025:ENSMUST00000160944 0.030098
ENSMUSG00000090025:ENSMUST00000160944 0.030098
. . .
0.007850
ENSMUSG00000090025:ENSMUST00000160944
0.007850
ENSMUSG00000090025:ENSMUST00000160944
0.021899
ENSMUSG00000090025:ENSMUST00000160944
0.021899
ENSMUSG00000090025:ENSMUST00000160944
0.030098
ENSMUSG00000090025:ENSMUST00000160944
0.030098
...
Finally, the computed offsets can be separated in genes of high versus
low expression, histogrammed using the vectors tool (operation 'hist')
and plotted using R (see Figure 8.5 for a sample plot), Excel or any other
similar tool or environment. For example, if the 'offset.txt' fi le computed
above was separated into two fi les 'offset.high.txt' for the genes of high
expression and 'offset.low.txt' for the genes of low expression, then:
$ cat offset.high.txt vectors -hist -n 6 -b 100 > profi le.high.txt
$ cat offset.high.txt vectors -hist -n 6 -b 100 > profi le.high.txt
$ cat offset.low.txt vectors -hist -n 6 -b 100 > profi le.low.txt
$ cat offset.low.txt vectors -hist -n 6 -b 100 > profi le.low.txt
$ head offset.high.txt
#bin-start
$ head offset.high.txt
#bin-start
bin-freq
bin-freq
bin-counts
bin-counts
0.000000
0.000000
0.008555
0.008555
20676
20676
0.010000
0.010000
0.008522
0.008522
20596
20596
0.020000
0.020000
0.008128
0.008128
19644
19644
. . .
. . .
Example of TSS read profi le for genes of high
and low expression
Figure 8.5






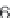
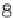




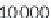

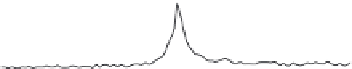




















Search WWH ::

Custom Search