Biology Reference
In-Depth Information
Bradford assay
Ninhydrin assay
4
4
BSA
BSA
3
3
Lysozyme
2
2
β
-Lactoglobulin
Lysozyme
1
1
β
-Lactoglobulin
0
0
0
5
10
15
20
0
5
10
15
20
Protein (
m
g/ml)
Protein (mg/ml)
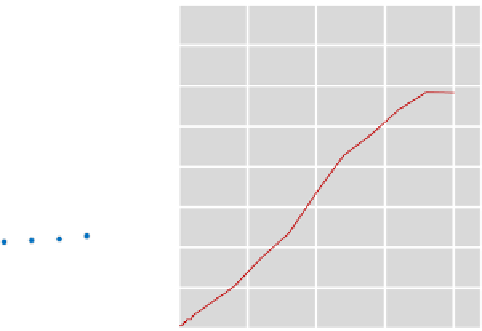
FIGURE 3.3
Evaluation of different assays for the determination of protein concentration. Absorption
curves have been determined for different standard proteins (BSA, bovine serum albumin;
b
-LG, beta-lactoglobulin; CSA, chicken serum albumin) with two different protein
concentration assays. These clearly show that the Ninhydrin-based protein assay
outperforms the Bradford assay in terms of linearity and lacking bias towards specific
proteins.
This means that, specific protein features aside, the primary sequence alone should
be used for the precise determination of protein concentration. The peptide bond
exactly fits this need. By using acid hydrolysis, the alpha amino groups can be
released from the peptide bonds and used as the reactive group to form the purple
coloured imino derivative of Ninhydrin. Photometric measurement of the purple
colour gives a proportional value of the amount of peptide bonds and, thereby, of
the amount of protein within the sample (
Figure 3.3
).
4
GENERATION OF ABSOLUTE QUANTITATIVE DATA
BY TARGETED MASS SPECTROMETRY
4.1
Criteria for selection of peptides for targeted analyses
One of the approaches to determining the absolute protein abundance in a proteome
sample is targeted analysis by SRM, relying on the stable isotope dilution concept.
The key practical consideration that needs to be considered for such analyses is which
proteins to target. This requires gathering as much information as possible from pre-
vious proteomic experiments about potential targets. Because the peptides used to rep-
resent the target proteins serve as surrogates for quantitation (
Kirkpatrick
et al.
, 2005
),