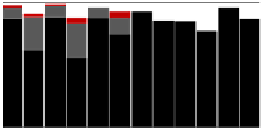Biology Reference
In-Depth Information
Complete
disruption
100
80
60
40
3rd cycle
2nd cylce
1st cycle
20
0
Growth phase
Duration disrupt. cycle
Type of disruptor
Organism
Exp Stat Exp Stat Exp Stat Exp Stat Exp Stat Exp Stat
Exp Stat Exp Stat Exp Stat Exp Stat Exp Stat Exp Stat
20s
30s
40s
60s
90s
120s
20s
30s
40s
180s
240s
300s
Ribolyser
Sonicator
Precellys
Dismembrator
B. subtilis
S. aureus
FIGURE 3.2
Evaluation of disruption efficiencies. Different cell lysis methods for the Gram-positive
bacteria
have been tested for both exponentially growing cells and
stationary phase cells. By using optimized lysis parameters, disruption efficiencies of
B. subtilis
and
S. aureus
90%
are achievable.
3.1.6
Determination of protein concentration
The determination of protein concentration is crucial in absolute quantitative
workflows. The failure to consider potential variations would lead to errors in
precision and accuracy that would be amplified in all downstream calculations aimed
at determining protein content.
Prior to any proteome analysis, it is strongly recommended to undertake an
evaluation of the existing methods for determining protein content. A vast number
of commercial systems exist that are based on the physical interactions or biochem-
ical properties of proteins. Although the choice for a specific system is likely to be
dependent on existing and widely available methods, consideration still needs to
be given to appropriateness of the chosen assay. Such considerations include the
presence of interfering chemicals and components in the buffers/media used, as well
as the average amino acid composition of the proteins under investigation. As shown
in
Figure 3.3
, we have compared a commercially available Bradford assay with an
assay that is based on the Ninhydrin reagent. The commonly used Bradford assay is
cheap, easy to use and sensitive. The active ingredient is Coomassie Brilliant Blue
G250, a disulfonylated triphenylmethane dye, that binds to proteins at positively
charged amino acids (arginine, lysine and histidine) via its sulfonyl groups and at
hydrophobic amino acids (tryptophan, tyrosine and phenylalanine) and aliphatic
chains of SDS within the protein via its triphenylmethane backbone (
Tal
et al.
,
1985
). This makes it sensitive to sequence variations, especially for amino acids that
are involved in the electrostatic and hydrophobic binding of the dye. The traditional
protein standard bovine serum albumin should not be used in connection with
Coomassie G250 because it has a much higher affinity for this dye compared to other
proteins.
The issue of biased protein content with respect to quantification can be circum-
vented by using detection agents that do not show specificity for sequence variations.