Biomedical Engineering Reference
In-Depth Information
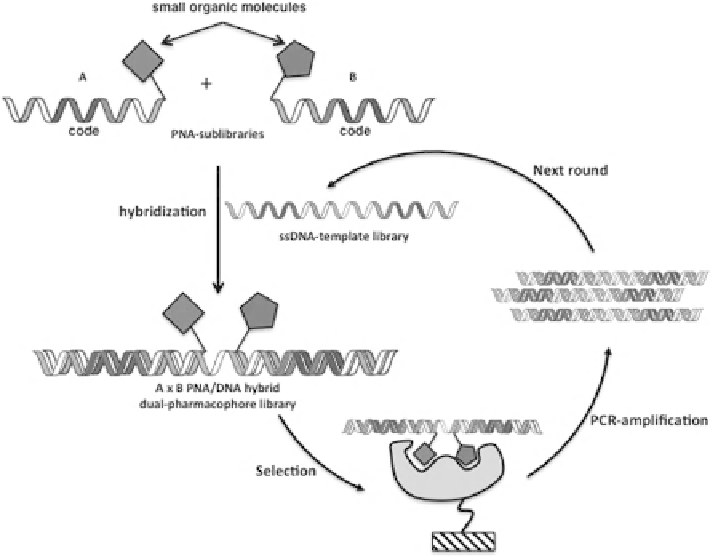
FIGURE 11.21
PNA/DNA combinatorial self-assembly encoded library. Two independent
PNA sublibraries, each displaying a chemical moiety and a compound-specific coding region,
are hybridized on a complementary DNA-template library. Following selection and PCR ampli-
fication of the DNA tags selected, dsDNA is converted into ssDNA. The second-generation
library is reassembled, exposing the templates to an excess of the fragment PNA sublibraries.
More recently, Winssinger's group has developed a dual-pharmacophore strategy
conceptually similar to ESAC technology, based on the combinatorial self-assembly
of two PNA libraries on single-stranded DNA templates (Figure 11.21) [99,100].
This procedure allows for the formation of a hybrid PNA/DNA structure which
encodes the chemical moieties in a manner compatible with PCR amplification
(Figure 11.21).
In principle, all the chemistry can be performed in an automated fashion (using a
common peptide synthesizer for the synthesis of the PNA conjugates and an oligonu-
cleotide synthesizer for the DNA templates) [100]. However, for the hypothetical
construction of a 1-million item compound library (1000
1000 library member),
it would require the laborious and expensive synthesis of 2000 PNA conjugates as
well as a tedious combinatorial enzymatic assembling of 1 million different DNA
templates (e.g., by mix and split of 2000 preconstituted DNA templates).
Nonetheless, the strategy presented by Daguer et al. lends itself to the molecular
evolution of its library members by the reannealing of PNA fragments after selection
×