Biomedical Engineering Reference
In-Depth Information
should be randomized to create the new binding surface.
Solvent accessibility and relative structural independence
of the residues located in the AB, CD, and EF loops were
judged based on the crystal structure of an Fc fragment of
human IgG1 (PDB code 1OQO). The degree of evolutionary
conservation of the residues was evaluated using the ConSurf
Server [3] and was taken as a further guide for selecting the
residues to be mutated. Fcab libraries were designed essen-
tially as described in Reference [2]. Together, the randomized
positions in these libraries yield a coherent surface of
5
4
3
HER-2/neu
TNF-
α
lgE
PE
2
1
0
-1
0
1
2
TNF355-2 log(ng/mL)
3
4
5
700-
1000A
2
, which compares well with the surface areas buried in
Fab-antigen interactions, which typically range from 600 to
900 A
2
[4]. A schematic presentation of the secondary struc-
ture of the CH3 domain [5], highlighting the randomized
positions, is given in Figure 39.2.
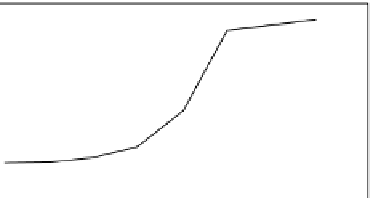
FIGURE 39.3
Antigen-specific binding of Fcab TNF353-2 to
immobilized TNF-
a
Bound Fcab was detected with antihuman
CH2 domain antibodies. PE, phycoerythrine.
facilitate exchange for the CH3 domain of Fcab TNF353-2.
HEK293 cells were transiently co-transfected with plasmids
encoding the TNF-
a
binding heavy chain and the light chain of
rituximab. After 5 days, supernatants were harvested and
mAb
2
protein RX-TNF353-2 was purified by Protein A
immuno-affinity chromatography. HPLC analysis showed
that the purified mAb
2
eluted as single peak at the expected
volume indistinguishable of the parental rituximab antibody.
To determine if the changes in the CH3 domain influenced
binding to Fc receptors known to be important for immuno-
effector functions (CD16a) and long in vivo half-life (FcRn),
surface plasmon resonance measurements were carried out.
At pH
39.4 TNF-a-BINDING Fcab: SELECTION AND
CHARACTERIZATION OF Fcab TNF353-2
For selection of an Fcab targeting the cytokine tumor necrosis
factor
a
(TNF-
a
), a Fcab library was generated in the yeast
expression vector pYD1 as described recently [2]. The result-
ing library expresses the
a
-agglutinin yeast adhesion receptor
Aga-2 as a fusion protein with Fcabs. Upon transformation of
the library into the Saccharomyces cerevisiae strain EBY100
(supplied transformed with the AGA1 gene), the Aga1 and the
Aga2-Fcab fusion protein associate within the secretory path-
way and upon translocation are displayed on the cell surface in
an inducible fashion [6]. Yeast cells expressing Fcabs on the
cell surface were incubated with biotinylated human TNF-
a
,
and cells interacting with the cytokine were enriched by high-
speed cell sorting essentially as described [2]. After five rounds
of sorting, the best TNF-
a
binders were screened by titrationof
individual yeast clones with biotinylated antigen. One of these
hits, Fcab TNF353-2, was chosen for further analysis. Fcab
TNF353-2 was expressed in mammalian cells as soluble Fcab
and purified by a single immuno-affinity chromatography step
using a Protein A column. The purified Fcab eluted as a single
peak from a size exclusion chromatography column at the
expected volume indicating the absence of protein aggregates,
fragments, or other contaminants. The purified Fcab bound
specifically to immobilized TNF-
a
in an enzyme-linked
immunosorbent assay (ELISA) with an EC
50
in the single
nano-molar range. This interaction appeared to be specific
since no binding to irrelevant antigens (IgE, HER2/neu, or
phycoerythrine) was detected (Figure 39.3).
For the generation of a prototypemAb
2
, the CH3 domain of
Fcab TNF353-2 was used to replace the CH3 domain of the
anti-CD20 specific antibody rituximab. This antibody is used
in the clinic for the treatment of blood malignancies [7]. The
heavy chain gene of rituximab was modified to create a silent
mutation leading to a unique XhoI restriction enzyme cleavage
site at the border between the CH2 and CH3 domain in order to
6, mAb
2
RX-TNF353-2 and its parent rituximab
(RX) bound with very similar affinities to human FcRn. At
pH
¼
7, both proteins completely lost their ability to interact
with the receptor indicating that the TNF-
a
binding site in the
mAb
2
did not interfere with the known pH-dependent FcRn
association and dissociation phenotype of normal antibodies.
Similar results were obtained when CD16a protein was
allowed to bind to the antibodies captured on Protein A
Biacore chips (Figure 39.4). These data suggested that the
in vivo half-life and immuno-effector functions of the original
rituximab antibody should be preserved in the mAb
2
.
It is known that rituximab is able to initiate immuno-
effector functions such as antibody-dependent cellular cyto-
toxicity (ADCC) and complement-dependent cytotoxicity
(CDC). To determine the ADCC potency of the mAb
2
purified primary human B lymphocytes were opsonized
with rituximab or the mAb
2
and then incubated with autol-
ogous natural killer (NK) cells as effector cells. After 4 h, the
killed B-cells were enumerated by flow cytometry after
staining dead cells with the fluorescent dye 7-amino-actino-
mycin D. The data demonstrated that both proteins mediated
NK cell dependent killing of B cells in a dose-dependent
manner and that the potencies of the mAb
2
and rituximab
were very similar (Figure 39.5A). No dead B cells were
measured in the absence of NK cells indicating that the
mechanism of cell death was indeed ADCC (not shown). To
measure CDC reactions, purified B cells were incubated
¼



















































Search WWH ::

Custom Search