Biology Reference
In-Depth Information
A-9
G-10.1
O
O
N
7
q
1
H
P
OH
O
Mg
2+
O
N
H
O
r
1
O
q
2
G-12
N
N
d
0
q
HA
NH
2
G-8
C-3
O
2
N
1
H
N
3
N
r
2
H
2
q
HB
q
3
O
-
r
8
r
HA
-
O
N
1
O
5
r
7
N
H
N
P
H
O
f
8
r
3
r
HB
O
2
q
inl
H
NH
2
H
O
q
7
O
C-1.1
N
O
C-17
N
N
N
H
N
O
O
N
OH
HN
O
O
r
5
r
4
G-2.1
H
H
q
5
q
4
H
N
H
N
H
2
N
N
r
6
N
N
O
H
N
N
G-5
A-14
q
6
N
N
N
N
N
Figure 2.10 Schematic representations of the HHR active sites and the two potentially
important hydrogen bond networks between C3 and G8 and between G5 and C17. All
key structural indexes calculated are also labeled.
5.1.1.1 Characterization of the active site structure and dynamics
of the WT simulation
The active site scaffold and hydrogen bond networks for the WT simulation
are depicted in
Fig. 2.10
. The eHHR ribozyme catalyzes the site-specific
cleavage by transesterification of the phosphodiester bond with a rate
enhancement up to 10
6
-fold relative to the rate of non-catalyzed cleavage.
39
The rate for cleavage and ligation for the naturally occurring eHHR motifs
are around 1000 and 2000 times faster, respectively, than the corresponding
rates for mHHR ribozymes.
84-86
Catalysis is generally believed to proceed by a general acid and base
mechanism. In this mechanism, the endocyclic amine of G12 (G12:N1)
in deprotonated form acts as the general base to abstract a proton from
the 2
0
OH of C17 (the nucleophile) to form an activated precursor. The acti-
vated precursor then proceeds by in-line attack on the adjacent scissile phos-
phate to form a pentacovalent phosphorane transition state. The 2
0
OH
group of G8 (G8:O2
0
) acts as a general acid catalyst to donate a proton to
the 5
0
oxygen of C1.1 (the leaving group) to facilitate breakdown of the



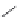







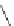



















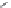














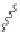





















































































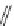































Search WWH ::

Custom Search