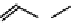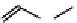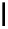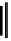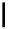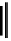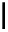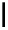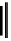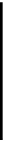Biology Reference
In-Depth Information
B
A
Replication
(passive
demethylation)
Passive or
active
demethylation ?
NH
2
NH
2
--CG--
--GC--
CH
3
N
N
N
N
O
O
C
5mC
Oxidation
(TET1/2/3)
NH
2
OH
Replication
--
C
G--
--GC--
--
C
G--
--G
C
--
--
C
G--
--G
C
--
CH
2
N
Maintenance
methylation
(DNMT1)
--
C
G--
--G
C
--
N
or
O
5hmC
C
= 5mC
C
= 5hmC
Figure 2.1 DNA methylation and hydroxymethylation occur on cytosines. (A) Chemical
structure of cytosine (C), 5-methylcytosine (5mC), and 5-hydroxymethylcytosine
(5hmC). (B) Cytosine methylation occurs symmetrically at CpG dinucleotides. De novo
methylation is mostly catalyzed by DNMT3A/B enzymes, whereas DNMT1 methylates
hemimethylated sites generated after DNA replication. Lack of maintenance methyla-
tion results in passive loss of DNA methylation with DNA replication. TET proteins are
capable of oxidizing 5mC into 5hmC, which acts as an intermediate in demethylation
pathways.
exciting prospect that 5hmC could also play major roles as an epigenetic
mark in development and disease, which prompted an impressive number
of studies in the past 4 years. In this review, we summarize the current
knowledge and discuss recent discoveries on the role of cytosine epigenetic
marks during mammalian development.
2. ENZYMATIC PLAYERS
2.1. DNA methyltransferases
The family of DNMTs comprises four members: DNMT1, DNMT3A,
DNMT3B, and DNMT3L (
Fig. 2.2
A). Another member initially identified
by sequence homologies, named DNMT2, was later found to methylate
RNA instead of DNA (
Goll et al., 2006
). DNMT1 is broadly expressed
and is responsible for propagating DNA methylation during cell division
by applying a methyl group on the newly synthesized DNA strand at
hemimethylated CpG sites created by DNA replication (
Jones & Liang,
2009
;
Fig. 2.1
B). This function of DNMT1 as a maintenance methyl-
transferase is supported by the fact that DNMT1 localizes at sites of DNA