Biomedical Engineering Reference
In-Depth Information
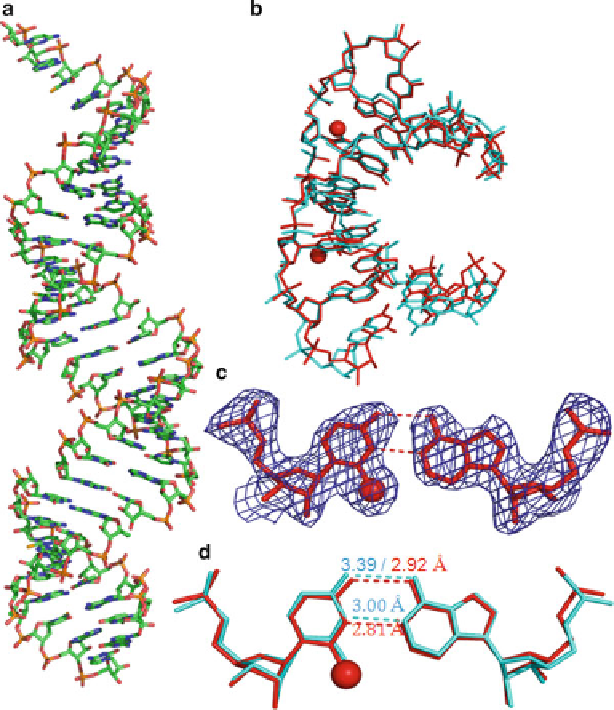
Fig. 3.7
Global and local structures of the
2-Se
U-containing RNA r[5
0
-GUAUA(
2-Se
U)AC-3
0
]
2
with a resolution of 2.3 A. (
a
) The overall structure of duplex. (
b
) The superimpose comparison
of one Se-U-RNA duplex (
red
; PDB ID: 3S49) with its native counterpart [5
0
-r(GUAUAUA)-
dC-3
0
]
2
(
cyan
; PDB ID: 246D) with an RMSD value of 0.55. The two
red
balls represent the
selenium atoms. (
c
) The experimental electron density of
2-Se
U6/A11 base pair with
1.0.
(
d
) The superimpose comparison of the local base pair
2-Se
U6/A11 (
red
)andthenative
2-Se
U6/A11
(
cyan
). The
numbers
indicate the distance between the corresponding atoms (Color figure online)
¢ D
can be converted to phosphoramidite and incorporated into oligonucleotides by
solid-phase synthesis under ultra-mild condition. The crystal structure (Fig.
3.8
)
of the 6-Se-dG-containing DNA and RNA duplex suggests the modified base-pair
shift by 0.3 A, while the modified
6-Se
G/C pairing is maintained. This observation
is consistent with the UV-melting experimental data, since the base-pair shift leads
to reduced stacking, thus decreasing the duplex stability. Moreover, the 6-Se-dG
modification in DNA also creates a characteristic UV absorption at 360 nm, which
Search WWH ::

Custom Search